Multiplex genome editing of microorganisms using CRISPR-Cas
- PMID: 31087001
- PMCID: PMC6522427
- DOI: 10.1093/femsle/fnz086
Multiplex genome editing of microorganisms using CRISPR-Cas
Abstract
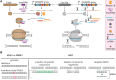
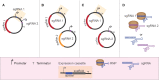
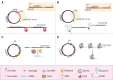
Microbial production of chemical compounds often requires highly engineered microbial cell factories. During the last years, CRISPR-Cas nucleases have been repurposed as powerful tools for genome editing. Here, we briefly review the most frequently used CRISPR-Cas tools and describe some of their applications. We describe the progress made with respect to CRISPR-based multiplex genome editing of industrial bacteria and eukaryotic microorganisms. We also review the state of the art in terms of gene expression regulation using CRISPRi and CRISPRa. Finally, we summarize the pillars for efficient multiplexed genome editing and present our view on future developments and applications of CRISPR-Cas tools for multiplex genome editing.
Keywords: CRISPR-Cas; Cas12a; Cas9; cell factories; genome editing; multiplex.
© FEMS 2019.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

