Genome-wide association study and genomic predictions for exterior traits in Yorkshire pigs1
- PMID: 31087081
- PMCID: PMC6606491
- DOI: 10.1093/jas/skz158
Genome-wide association study and genomic predictions for exterior traits in Yorkshire pigs1
Abstract
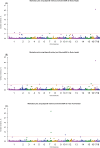
The objectives of this study were to identify informative genomic regions that affect the exterior traits of purebred Korean Yorkshire pigs and to investigate and compare the accuracy of genomic prediction for response variables. Phenotypic data on body height (BH), body length (BL), and total teat number (TTN) from 2,432 Yorkshire pigs were used to obtain breeding values including as response variable the estimated breeding value (EBV) and 2 types of deregressed EBVs-one including the parent average (DEBVincPA) and the other excluding it (DEBVexcPA). A final genotype panel comprising 46,199 SNP markers was retained for analysis after quality control for common SNPs. The BayesB and BayesC methods-with various π and weighted response variables (EBV, DEBVincPA, or DEBVexcPA)-were used to estimate SNP effects, through the genome-wide association study. The significance of genomic windows (1 Mb) was obtained at 1.0% additive genetic variance and was subsequently used to identify informative genomic regions. Furthermore, SNPs with a high model frequency (≥0.90) were considered informative. The accuracy of genomic prediction was estimated using a 5-fold cross-validation with the K-means clustering method. Genomic accuracy was measured as the genomic correlation between the molecular breeding value and the individual weighted response variables (EBV, DEBVincPA, or DEBVexcPA). The number of identified informative windows (1 Mb) for BH, BL, and TTN was 4, 3, and 4, respectively. The number of significant SNPs for BH, BL, and TTN was 6, 4, and 5, respectively. Diversity π did not influence the accuracy of genomic prediction. The BayesB method showed slightly higher genomic accuracy for exterior traits than BayesC method in this study. In addition, the genomic accuracy using DEBVincPA as response variable was higher than that using other response variables. Therefore, the genomic accuracy using BayesB (π = 0.90) with DEBVinPA as a response variable was the most effective in this study. The genomic accuracy values for BH, BL, and TTN were calculated to be 0.52, 0.60, and 0.51, respectively.
Keywords: Bayesian method; GWAS; Yorkshire pigs; exterior traits; genomic prediction.
© The Author(s) 2019. Published by Oxford University Press on behalf of the American Society of Animal Science. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Aguilar I., Misztal I., Tsuruta S., Legarra A., and Wang H.. 2014. PREGSF90–POSTGSF90: Computational tools for the implementation of single-step genomic selection and genome-wide association with ungenotyped individuals in BLUPF90 programs. In: Proc. 10th World Congr. Genet. Appl. Livest. Prod. 23 August, Canada. doi:10.13140/2.1.4801.5045
-
- Fernández A. I., Pérez-Montarelo D., Barragán C., Ramayo-Caldas Y., Ibáñez-Escriche N., Castelló A., Noguera J. L., Silió L., Folch J. M., and Rodríguez M. C.. 2012. Genome-wide linkage analysis of QTL for growth and body composition employing the Porcine SNP60 BeadChip. BMC Genet. 13:41. doi:10.1186/1471-2156-13-41 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

