The endoplasmic reticulum chaperone PfGRP170 is essential for asexual development and is linked to stress response in malaria parasites
- PMID: 31087747
- PMCID: PMC6699899
- DOI: 10.1111/cmi.13042
The endoplasmic reticulum chaperone PfGRP170 is essential for asexual development and is linked to stress response in malaria parasites
Abstract
The vast majority of malaria mortality is attributed to one parasite species: Plasmodium falciparum. Asexual replication of the parasite within the red blood cell is responsible for the pathology of the disease. In Plasmodium, the endoplasmic reticulum (ER) is a central hub for protein folding and trafficking as well as stress response pathways. In this study, we tested the role of an uncharacterised ER protein, PfGRP170, in regulating these key functions by generating conditional mutants. Our data show that PfGRP170 localises to the ER and is essential for asexual growth, specifically required for proper development of schizonts. PfGRP170 is essential for surviving heat shock, suggesting a critical role in cellular stress response. The data demonstrate that PfGRP170 interacts with the Plasmodium orthologue of the ER chaperone, BiP. Finally, we found that loss of PfGRP170 function leads to the activation of the Plasmodium eIF2α kinase, PK4, suggesting a specific role for this protein in this parasite stress response pathway.
Keywords: Plasmodium falciparum; endoplasmic reticulum; malaria; parasitology.
© 2019 The Authors Cellular Microbiology Published by John Wiley & Sons Ltd.
Figures
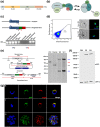
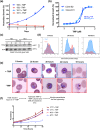


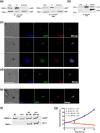
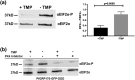
References
-
- Andreasson, C. , Rampelt, H. , Fiaux, J. , Druffel‐Augustin, S. , & Bukau, B. (2010). The endoplasmic reticulum Grp170 acts as a nucleotide exchange factor of Hsp70 via a mechanism similar to that of the cytosolic Hsp110. The Journal of Biological Chemistry, 285, 12445–12453. 10.1074/jbc.M109.096735 - DOI - PMC - PubMed
-
- Babbitt, S. E. , Altenhofen, L. , Cobbold, S. A. , Istvan, E. S. , Fennell, C. , Doerig, C. , … Goldberg, D. E. (2012). Plasmodium falciparum responds to amino acid starvation by entering into a hibernatory state. Proceedings of the National Academy of Sciences of the United States of America, 109, E3278–E3287. 10.1073/pnas.1209823109 - DOI - PMC - PubMed
-
- Balu, B. , Shoue, D. A. , Fraser, M. J. Jr. , & Adams, J. H. (2005). High‐efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proceedings of the National Academy of Sciences of the United States of America, 102, 16391–16396. 10.1073/pnas.0504679102 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 18POST34080315/American Heart Association Postdoctoral Fellowship/International
- T32 AI060546/AI/NIAID NIH HHS/United States
- T32AI060546/National Institute of Allergy and Infectious Diseases/International
- R00AI099156/National Institute of Allergy and Infectious Diseases/International
- R01 AI130139/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources

