The solution structure of the human IgG2 subclass is distinct from those for human IgG1 and IgG4 providing an explanation for their discrete functions
- PMID: 31088911
- PMCID: PMC6635440
- DOI: 10.1074/jbc.RA118.007134
The solution structure of the human IgG2 subclass is distinct from those for human IgG1 and IgG4 providing an explanation for their discrete functions
Abstract
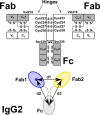
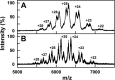
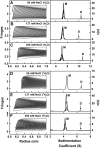
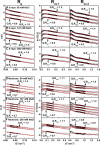
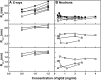
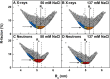
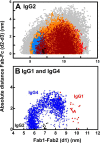
Human IgG2 antibody displays distinct therapeutically-useful properties compared with the IgG1, IgG3, and IgG4 antibody subclasses. IgG2 is the second most abundant IgG subclass, being able to bind human FcγRII/FcγRIII but not to FcγRI or complement C1q. Structural information on IgG2 is limited by the absence of a full-length crystal structure for this. To this end, we determined the solution structure of human myeloma IgG2 by atomistic X-ray and neutron-scattering modeling. Analytical ultracentrifugation disclosed that IgG2 is monomeric with a sedimentation coefficient (s20, w0) of 7.2 S. IgG2 dimer formation was ≤5% and independent of the buffer conditions. Small-angle X-ray scattering in a range of NaCl concentrations and in light and heavy water revealed that the X-ray radius of gyration (Rg ) is 5.2-5.4 nm, after allowing for radiation damage at higher concentrations, and that the neutron Rg value of 5.0 nm remained unchanged in all conditions. The X-ray and neutron distance distribution curves (P(r)) revealed two peaks, M1 and M2, that were unchanged in different buffers. The creation of >123,000 physically-realistic atomistic models by Monte Carlo simulations for joint X-ray and neutron-scattering curve fits, constrained by the requirement of correct disulfide bridges in the hinge, resulted in the determination of symmetric Y-shaped IgG2 structures. These molecular structures were distinct from those for asymmetric IgG1 and asymmetric and symmetric IgG4 and were attributable to the four hinge disulfides. Our IgG2 structures rationalize the existence of the human IgG1, IgG2, and IgG4 subclasses and explain the receptor-binding functions of IgG2.
Keywords: analytical ultracentrifugation; antibody; immunoglobulin G (IgG); molecular modeling; neutron-scattering; small-angle X-ray scattering (SAXS).
© 2019 Hui et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures






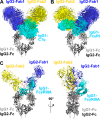
References
-
- Strohl W. R., and Strohl L. M. (eds) (2012) in Therapeutic Antibody Engineering, pp. 37–595, Woodhead Publishing Series in Biomedicine, Woodhead Publishing, Sawston, Cambridge, UK
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

