Transient self-organisation of DNA coated colloids directed by enzymatic reactions
- PMID: 31089164
- PMCID: PMC6517385
- DOI: 10.1038/s41598-019-43720-7
Transient self-organisation of DNA coated colloids directed by enzymatic reactions
Abstract
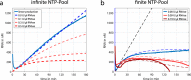
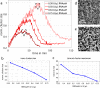
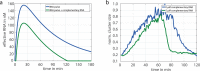
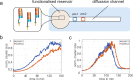
Dynamic self-organisation far from equilibrium is a key concept towards building autonomously acting materials. Here, we report the coupling of an antagonistic enzymatic reaction of RNA polymerisation and degradation to the aggregation of micron sized DNA coated colloids into fractal structures. A transient colloidal aggregation process is controlled by competing reactions of RNA synthesis of linker strands by a RNA polymerase and their degradation by a ribonuclease. By limiting the energy supply (NTP) of the enzymatic reactions, colloidal clusters form and subsequently disintegrate without the need of external stimuli. Here, the autonomous colloidal aggregation and disintegration can be modulated in terms of lifetime and cluster size. By restricting the enzyme activity locally, a directed spatial propagation of a colloidal aggregation and disintegration front is realised.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Rueb CJ, Zukoski CF. Viscoelastic properties of colloidal gels. Journal of Rheology. 1997;41:197–218. doi: 10.1122/1.550812. - DOI
-
- Shih, W.-H. et al. Mechanical Properties of Colloidal Gels. MRS Proc. 155 (1989).
-
- Kamp SW, Kilfoil ML. Universal behaviour in the mechanical properties of weakly aggregated colloidal particles. Soft matter. 2009;5:2438. doi: 10.1039/b814975e. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

