Acetate Promotes T Cell Effector Function during Glucose Restriction
- PMID: 31091446
- PMCID: PMC6544383
- DOI: 10.1016/j.celrep.2019.04.022
Acetate Promotes T Cell Effector Function during Glucose Restriction
Abstract
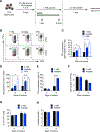
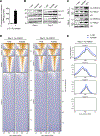
Competition for nutrients like glucose can metabolically restrict T cells and contribute to their hyporesponsiveness during cancer. Metabolic adaptation to the surrounding microenvironment is therefore key for maintaining appropriate cell function. For instance, cancer cells use acetate as a substrate alternative to glucose to fuel metabolism and growth. Here, we show that acetate rescues effector function in glucose-restricted CD8+ T cells. Mechanistically, acetate promotes histone acetylation and chromatin accessibility and enhances IFN-γ gene transcription and cytokine production in an acetyl-CoA synthetase (ACSS)-dependent manner. Ex vivo acetate treatment increases IFN-γ production by exhausted T cells, whereas reducing ACSS expression in T cells impairs IFN-γ production by tumor-infiltrating lymphocytes and tumor clearance. Thus, hyporesponsive T cells can be epigenetically remodeled and reactivated by acetate, suggesting that pathways regulating the use of substrates alternative to glucose could be therapeutically targeted to promote T cell function during cancer.
Keywords: T cell exhaustion; T cell hyporesponsiveness; T cells; acetate; acetyl-CoA synthetase; chromatin remodeling; effector functions; tumor immunity; tumor-infiltrating lymphocytes.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
E.J.P. is a founder of Rheos Medicines and E.L.P. is an SAB member of ImmunoMet and a founder of Rheos Medicines.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

