MicroRNA-615-5p Regulates Angiogenesis and Tissue Repair by Targeting AKT/eNOS (Protein Kinase B/Endothelial Nitric Oxide Synthase) Signaling in Endothelial Cells
- PMID: 31092013
- PMCID: PMC6594892
- DOI: 10.1161/ATVBAHA.119.312726
MicroRNA-615-5p Regulates Angiogenesis and Tissue Repair by Targeting AKT/eNOS (Protein Kinase B/Endothelial Nitric Oxide Synthase) Signaling in Endothelial Cells
Abstract
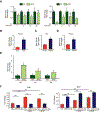
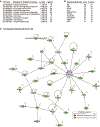
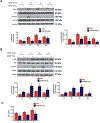
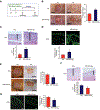
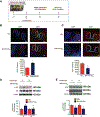
Objective- In response to tissue injury, the appropriate progression of events in angiogenesis is controlled by a careful balance between pro and antiangiogenic factors. We aimed to identify and characterize microRNAs that regulate angiogenesis in response to tissue injury. Approach and Results- We show that in response to tissue injury, microRNA-615-5p (miR-615-5p) is rapidly induced and serves as an antiangiogenic microRNA by targeting endothelial cell VEGF (vascular endothelial growth factor)-AKT (protein kinase B)/eNOS (endothelial nitric oxide synthase) signaling in vitro and in vivo. MiR-615-5p expression is increased in wounds of diabetic db/db mice, in plasma of human subjects with acute coronary syndromes, and in plasma and skin of human subjects with diabetes mellitus. Ectopic expression of miR-615-5p markedly inhibited endothelial cell proliferation, migration, network tube formation in Matrigel, and the release of nitric oxide, whereas miR-615-5p neutralization had the opposite effects. Mechanistic studies using transcriptomic profiling, bioinformatics, 3' untranslated region reporter and microribonucleoprotein immunoprecipitation assays, and small interfering RNA dependency studies demonstrate that miR-615-5p inhibits the VEGF-AKT/eNOS signaling pathway in endothelial cells by targeting IGF2 (insulin-like growth factor 2) and RASSF2 (Ras-associating domain family member 2). Local delivery of miR-615-5p inhibitors, markedly increased angiogenesis, granulation tissue thickness, and wound closure rates in db/db mice, whereas miR-615-5p mimics impaired these effects. Systemic miR-615-5p neutralization improved skeletal muscle perfusion and angiogenesis after hindlimb ischemia in db/db mice. Finally, modulation of miR-615-5p expression dynamically regulated VEGF-induced AKT signaling and angiogenesis in human skin organoids as a model of tissue injury. Conclusions- These findings establish miR-615-5p as an inhibitor of VEGF-AKT/eNOS-mediated endothelial cell angiogenic responses and that manipulating miR-615-5p expression could provide a new target for angiogenic therapy in response to tissue injury. Visual Overview- An online visual overview is available for this article.
Keywords: AKT/eNOS signaling; angiogenesis; endothelial cells; microRNAs; tissue repair.
Conflict of interest statement
Disclosures
The authors have declared that no conflict of interest exists.
Figures



Similar articles
-
MicroRNA-135a-3p regulates angiogenesis and tissue repair by targeting p38 signaling in endothelial cells.FASEB J. 2019 Apr;33(4):5599-5614. doi: 10.1096/fj.201802063RR. Epub 2019 Jan 22. FASEB J. 2019. PMID: 30668922 Free PMC article.
-
MicroRNA-199a-3p and MicroRNA-199a-5p Take Part to a Redundant Network of Regulation of the NOS (NO Synthase)/NO Pathway in the Endothelium.Arterioscler Thromb Vasc Biol. 2018 Oct;38(10):2345-2357. doi: 10.1161/ATVBAHA.118.311145. Arterioscler Thromb Vasc Biol. 2018. PMID: 29976767
-
miR-125a-5p impairs endothelial cell angiogenesis in aging mice via RTEF-1 downregulation.Aging Cell. 2014 Oct;13(5):926-34. doi: 10.1111/acel.12252. Epub 2014 Jul 24. Aging Cell. 2014. PMID: 25059272 Free PMC article.
-
AngiomiRs--key regulators of angiogenesis.Curr Opin Genet Dev. 2009 Jun;19(3):205-11. doi: 10.1016/j.gde.2009.04.002. Epub 2009 May 14. Curr Opin Genet Dev. 2009. PMID: 19446450 Free PMC article. Review.
-
The potential of miR-29 in modulating tumor angiogenesis: a comprehensive review.Discov Oncol. 2025 Apr 6;16(1):474. doi: 10.1007/s12672-025-02246-3. Discov Oncol. 2025. PMID: 40189720 Free PMC article. Review.
Cited by
-
Gene therapy to enhance angiogenesis in chronic wounds.Mol Ther Nucleic Acids. 2022 Aug 17;29:871-899. doi: 10.1016/j.omtn.2022.08.020. eCollection 2022 Sep 13. Mol Ther Nucleic Acids. 2022. PMID: 36159590 Free PMC article. Review.
-
MicroRNAomic Analysis of Spent Media from Slow- and Fast-Growing Bovine Embryos Reveal Distinct Differences.Animals (Basel). 2024 Aug 13;14(16):2331. doi: 10.3390/ani14162331. Animals (Basel). 2024. PMID: 39199865 Free PMC article.
-
miR-615 Fine-Tunes Growth and Development and Has a Role in Cancer and in Neural Repair.Cells. 2020 Jun 27;9(7):1566. doi: 10.3390/cells9071566. Cells. 2020. PMID: 32605009 Free PMC article. Review.
-
Nicotinamide mononucleotide (NMN) protects bEnd.3 cells against H2 O2 -induced damage via NAMPT and the NF-κB p65 signalling pathway.FEBS Open Bio. 2021 Mar;11(3):866-879. doi: 10.1002/2211-5463.13067. Epub 2021 Feb 9. FEBS Open Bio. 2021. PMID: 33340447 Free PMC article.
-
Extracellular Vesicle miRNAs in the Promotion of Cardiac Neovascularisation.Front Physiol. 2020 Sep 25;11:579892. doi: 10.3389/fphys.2020.579892. eCollection 2020. Front Physiol. 2020. PMID: 33101061 Free PMC article. Review.
References
-
- Blakytny R, Jude E. The molecular biology of chronic wounds and delayed healing in diabetes. Diabet Med. 2006;23:594–608 - PubMed
-
- Falanga V Wound healing and its impairment in the diabetic foot. Lancet. 2005;366:1736–1743 - PubMed
-
- Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

