Proteome of the central apparatus of a ciliary axoneme
- PMID: 31092556
- PMCID: PMC6548120
- DOI: 10.1083/jcb.201902017
Proteome of the central apparatus of a ciliary axoneme
Abstract
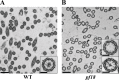
Nearly all motile cilia have a "9+2" axoneme containing a central apparatus (CA), consisting of two central microtubules with projections, that is essential for motility. To date, only 22 proteins are known to be CA components. To identify new candidate CA proteins, we used mass spectrometry to compare axonemes of wild-type Chlamydomonas and a CA-less mutant. We identified 44 novel candidate CA proteins, of which 13 are conserved in humans. Five of the latter were studied more closely, and all five localized to the CA; therefore, most of the other candidates are likely to also be CA components. Our results reveal that the CA is far more compositionally complex than previously recognized and provide a greatly expanded knowledge base for studies to understand the architecture of the CA and how it functions. The discovery of the new conserved CA proteins will facilitate genetic screening to identify patients with a form of primary ciliary dyskinesia that has been difficult to diagnose.
© 2019 Zhao et al.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

