Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells
- PMID: 31092822
- PMCID: PMC6520360
- DOI: 10.1038/s41467-019-10122-2
Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells
Abstract
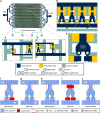
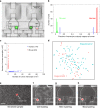
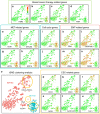
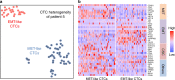
Molecular analysis of circulating tumor cells (CTCs) at single-cell resolution offers great promise for cancer diagnostics and therapeutics from simple liquid biopsy. Recent development of massively parallel single-cell RNA-sequencing (scRNA-seq) provides a powerful method to resolve the cellular heterogeneity from gene expression and pathway regulation analysis. However, the scarcity of CTCs and the massive contamination of blood cells limit the utility of currently available technologies. Here, we present Hydro-Seq, a scalable hydrodynamic scRNA-seq barcoding technique, for high-throughput CTC analysis. High cell-capture efficiency and contamination removal capability of Hydro-Seq enables successful scRNA-seq of 666 CTCs from 21 breast cancer patient samples at high throughput. We identify breast cancer drug targets for hormone and targeted therapies and tracked individual cells that express markers of cancer stem cells (CSCs) as well as of epithelial/mesenchymal cell state transitions. Transcriptome analysis of these cells provides insights into monitoring target therapeutics and processes underlying tumor metastasis.
Conflict of interest statement
E.Y., Y.-H.C., Y.-C.C., and R.B. filed a patent application on the Hydro-Seq technology (Application No.: 62/449,867 and PCT/US2018/014353). S.N., E.L., and M.S.W. have a patent (US20150293010A1) on the Labyrinth enrichment technology. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- R35 CA 129765/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- R01 CA 203810/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
- R01 CA203810/CA/NCI NIH HHS/United States
- 2017 Translational Research Partnership Program/University of Michigan (U-M)/International
- R21 CA 195016/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

