Biomimetic electromechanical stimulation to maintain adult myocardial slices in vitro
- PMID: 31092830
- PMCID: PMC6520377
- DOI: 10.1038/s41467-019-10175-3
Biomimetic electromechanical stimulation to maintain adult myocardial slices in vitro
Abstract
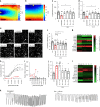
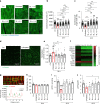
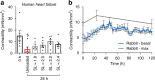
Adult cardiac tissue undergoes a rapid process of dedifferentiation when cultured outside the body. The in vivo environment, particularly constant electromechanical stimulation, is fundamental to the regulation of cardiac structure and function. We investigated the role of electromechanical stimulation in preventing culture-induced dedifferentiation of adult cardiac tissue using rat, rabbit and human heart failure myocardial slices. Here we report that the application of a preload equivalent to sarcomere length (SL) = 2.2 μm is optimal for the maintenance of rat myocardial slice structural, functional and transcriptional properties at 24 h. Gene sets associated with the preservation of structure and function are activated, while gene sets involved in dedifferentiation are suppressed. The maximum contractility of human heart failure myocardial slices at 24 h is also optimally maintained at SL = 2.2 μm. Rabbit myocardial slices cultured at SL = 2.2 μm remain stable for 5 days. This approach substantially prolongs the culture of adult cardiac tissue in vitro.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
- RM/17/1/33377/BHF_/British Heart Foundation/United Kingdom
- MR/L012618/1/MRC_/Medical Research Council/United Kingdom
- FS/16/17/31663/BHF_/British Heart Foundation/United Kingdom
- RM/17/1/33377/British Heart Foundation (BHF)/International
- MR/R010676/1/RCUK | Medical Research Council (MRC)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

