Tracing the origin of adult intestinal stem cells
- PMID: 31092921
- PMCID: PMC6986928
- DOI: 10.1038/s41586-019-1212-5
Tracing the origin of adult intestinal stem cells
Abstract
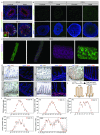
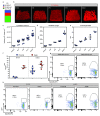
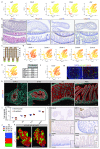
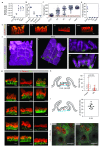
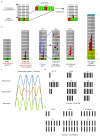
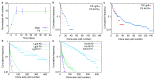
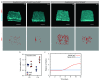
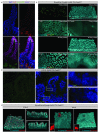
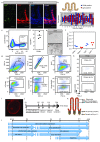
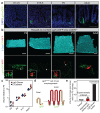
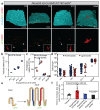
Adult intestinal stem cells are located at the bottom of crypts of Lieberkühn, where they express markers such as LGR51,2 and fuel the constant replenishment of the intestinal epithelium1. Although fetal LGR5-expressing cells can give rise to adult intestinal stem cells3,4, it remains unclear whether this population in the patterned epithelium represents unique intestinal stem-cell precursors. Here we show, using unbiased quantitative lineage-tracing approaches, biophysical modelling and intestinal transplantation, that all cells of the mouse intestinal epithelium-irrespective of their location and pattern of LGR5 expression in the fetal gut tube-contribute actively to the adult intestinal stem cell pool. Using 3D imaging, we find that during fetal development the villus undergoes gross remodelling and fission. This brings epithelial cells from the non-proliferative villus into the proliferative intervillus region, which enables them to contribute to the adult stem-cell niche. Our results demonstrate that large-scale remodelling of the intestinal wall and cell-fate specification are closely linked. Moreover, these findings provide a direct link between the observed plasticity and cellular reprogramming of differentiating cells in adult tissues following damage5-9, revealing that stem-cell identity is an induced rather than a hardwired property.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Comment in
-
Origins of intestinal stem cells - all in it together?Nat Rev Gastroenterol Hepatol. 2019 Jul;16(7):389. doi: 10.1038/s41575-019-0166-2. Nat Rev Gastroenterol Hepatol. 2019. PMID: 31164775 No abstract available.
References
-
- Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

