Identifying Clinical Terms in Medical Text Using Ontology-Guided Machine Learning
- PMID: 31094361
- PMCID: PMC6533869
- DOI: 10.2196/12596
Identifying Clinical Terms in Medical Text Using Ontology-Guided Machine Learning
Abstract
Background: Automatic recognition of medical concepts in unstructured text is an important component of many clinical and research applications, and its accuracy has a large impact on electronic health record analysis. The mining of medical concepts is complicated by the broad use of synonyms and nonstandard terms in medical documents.
Objective: We present a machine learning model for concept recognition in large unstructured text, which optimizes the use of ontological structures and can identify previously unobserved synonyms for concepts in the ontology.
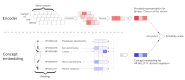
Methods: We present a neural dictionary model that can be used to predict if a phrase is synonymous to a concept in a reference ontology. Our model, called the Neural Concept Recognizer (NCR), uses a convolutional neural network to encode input phrases and then rank medical concepts based on the similarity in that space. It uses the hierarchical structure provided by the biomedical ontology as an implicit prior embedding to better learn embedding of various terms. We trained our model on two biomedical ontologies-the Human Phenotype Ontology (HPO) and Systematized Nomenclature of Medicine - Clinical Terms (SNOMED-CT).
Results: We tested our model trained on HPO by using two different data sets: 288 annotated PubMed abstracts and 39 clinical reports. We achieved 1.7%-3% higher F1-scores than those for our strongest manually engineered rule-based baselines (P=.003). We also tested our model trained on the SNOMED-CT by using 2000 Intensive Care Unit discharge summaries from MIMIC (Multiparameter Intelligent Monitoring in Intensive Care) and achieved 0.9%-1.3% higher F1-scores than those of our baseline. The results of our experiments show high accuracy of our model as well as the value of using the taxonomy structure of the ontology in concept recognition.
Conclusion: Most popular medical concept recognizers rely on rule-based models, which cannot generalize well to unseen synonyms. In addition, most machine learning methods typically require large corpora of annotated text that cover all classes of concepts, which can be extremely difficult to obtain for biomedical ontologies. Without relying on large-scale labeled training data or requiring any custom training, our model can be efficiently generalized to new synonyms and performs as well or better than state-of-the-art methods custom built for specific ontologies.
Keywords: biomedical ontologies; concept recognition; human phenotype ontology; machine learning; medical text mining; phenotyping.
©Aryan Arbabi, David R Adams, Sanja Fidler, Michael Brudno. Originally published in JMIR Medical Informatics (http://medinform.jmir.org), 10.05.2019.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures

References
-
- Simmons Michael, Singhal Ayush, Lu Zhiyong. Text Mining for Precision Medicine: Bringing Structure to EHRs and Biomedical Literature to Understand Genes and Health. Adv Exp Med Biol. 2016;939:139–166. doi: 10.1007/978-981-10-1503-8_7. http://europepmc.org/abstract/MED/27807747 - DOI - PMC - PubMed
-
- Jonnagaddala H. Healthcare Ethics and Training: Concepts, Methodologies, Tools, and Applications. PA, USA: IGI Global; 2017. Mining Electronic Health Records to Guide and Support Clinical Decision Support Systems; pp. 184–201.
-
- Gonzalez GH, Tahsin T, Goodale BC, Greene AC, Greene CS. Recent Advances and Emerging Applications in Text and Data Mining for Biomedical Discovery. Brief Bioinform. 2016 Jan;17(1):33–42. doi: 10.1093/bib/bbv087. http://europepmc.org/abstract/MED/26420781 bbv087 - DOI - PMC - PubMed
-
- Piñero J, Queralt-Rosinach N, Bravo A, Deu-Pons J, Bauer-Mehren A, Baron M, Sanz F, Furlong LI. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database (Oxford) 2015;2015:bav028. doi: 10.1093/database/bav028. http://europepmc.org/abstract/MED/25877637 bav028 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

