An Ontology for Facilitating Discussion of Catalytic Strategies of RNA-Cleaving Enzymes
- PMID: 31095369
- PMCID: PMC6661149
- DOI: 10.1021/acschembio.9b00202
An Ontology for Facilitating Discussion of Catalytic Strategies of RNA-Cleaving Enzymes
Abstract
A predictive understanding of the mechanisms of RNA cleavage is important for the design of emerging technology built from biological and synthetic molecules that have promise for new biochemical and medicinal applications. Over the past 15 years, RNA cleavage reactions involving 2'-O-transphosphorylation have been discussed using a simplified framework introduced by Breaker that consists of four fundamental catalytic strategies (designated α, β, γ, and δ) that contribute to rate enhancement. As more detailed mechanistic data emerge, there is need for the framework to evolve and keep pace. We develop an ontology for discussion of strategies of enzymes that catalyze RNA cleavage via 2'-O-transphosphorylation that stratifies Breaker's framework into primary (1°), secondary (2°), and tertiary (3°) contributions to enable more precise interpretation of mechanism in the context of structure and bonding. Further, we point out instances where atomic-level changes give rise to changes in more than one catalytic contribution, a phenomenon we refer to as "functional blurring". We hope that this ontology will help clarify our conversations and pave the path forward toward a consensus view of these fundamental and fascinating mechanisms. The insight gained will deepen our understanding of RNA cleavage reactions catalyzed by natural protein and RNA enzymes, as well as aid in the design of new engineered DNA and synthetic enzymes.
Figures
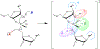
 - Arrangement of the O2′ nucleophile, P (scissile phosphorus), and O5′ leaving group in an in-line attack geometry (facilitated by contacts, indicated by blue arcs, that splay the N−1 and N+1 bases).
- Arrangement of the O2′ nucleophile, P (scissile phosphorus), and O5′ leaving group in an in-line attack geometry (facilitated by contacts, indicated by blue arcs, that splay the N−1 and N+1 bases).  - Stabilization (neutralization/protonation) of the negative charge accumulation on the non-bridging phosphoryl oxygens (NPOs).
- Stabilization (neutralization/protonation) of the negative charge accumulation on the non-bridging phosphoryl oxygens (NPOs).  - Activation (deprotonation) of the O2′ nucleophile.
- Activation (deprotonation) of the O2′ nucleophile.  - Stabilization (neutralization/protonation) of the accumulating negative charge on the O5′ leaving group. Although this schematic uses a transition state model to illustrate the fundamental catalytic strategies, these strategies can impact any state along the reaction coordinate. Colored ovals highlight each strategy and encompass the primary atomic positions (defined in section 4 below) associated with the chemical space of bonds for each strategy.
- Stabilization (neutralization/protonation) of the accumulating negative charge on the O5′ leaving group. Although this schematic uses a transition state model to illustrate the fundamental catalytic strategies, these strategies can impact any state along the reaction coordinate. Colored ovals highlight each strategy and encompass the primary atomic positions (defined in section 4 below) associated with the chemical space of bonds for each strategy.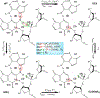
References
-
- Perreault DM; Anslyn EV, Unifying the current data on the mechanism of cleavage– transesterification of RNA. Angew. Chem., Int. Ed 1997, 36 (5), 432–450.
-
- Oivanen M; Kuusela S; Lönnberg H, Kinetics and mechanisms for the cleavage and isomerization of the phosphodiester bonds of RNA by Brønsted acids and bases. Chem. Rev 1998, 98 (3), 961–990. - PubMed
-
- Korhonen H; Williams NH; Mikkola S, βLG values in mechanistic studies on the transesterification of RNA models and their application in a metal ion complex promoted transesterification. J. Phys. Org. Chem 2013, 26 (2), 182–186.
-
- Lilley DM, How RNA acts as a nuclease: some mechanistic comparisons in the nucleolytic ribozymes. Biochem. Soc. Trans 2017, 45 (3), 683–691. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

