Topological Metrizations of Trees, and New Quartet Methods of Tree Inference
- PMID: 31095496
- PMCID: PMC7650847
- DOI: 10.1109/TCBB.2019.2917204
Topological Metrizations of Trees, and New Quartet Methods of Tree Inference
Abstract
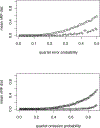
Topological phylogenetic trees can be assigned edge weights in several natural ways, highlighting different aspects of the tree. Here, the rooted triple and quartet metrizations are introduced, and applied to formulate novel methods of inferring large trees from rooted triple and quartet data. These methods lead to new statistically consistent procedures for inference of a species tree from gene trees under the multispecies coalescent model.
Figures




References
-
- Allman ES, Degnan JH, and Rhodes JA Identifying the rooted species tree from the distribution of unrooted gene trees under the coalescent. J. Math. Biol, 62(6):833–862, 2011. - PubMed
-
- Allman ES, Degnan JH, and Rhodes JA Species tree inference by the STAR method, and generalizations. J. Comput. Biol, 20(1):50–61, 2013. - PubMed
-
- Anvi E, Cohen R, and Snir S Weighted quartets phylogenetics. Syst. Biol, 64(2):233–242, 2015. - PubMed
-
- Bandelt H-J and Dress A Reconstructing the shape of a tree from observed dissimilarity data. Adv. Appl. Math, 7:309–343, 1986.

