Computational identification of tissue-specific transcription factor cooperation in ten cattle tissues
- PMID: 31095599
- PMCID: PMC6522001
- DOI: 10.1371/journal.pone.0216475
Computational identification of tissue-specific transcription factor cooperation in ten cattle tissues
Abstract
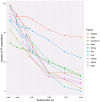
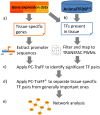
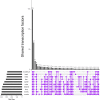
Transcription factors (TFs) are a special class of DNA-binding proteins that orchestrate gene transcription by recruiting other TFs, co-activators or co-repressors. Their combinatorial interplay in higher organisms maintains homeostasis and governs cell identity by finely controlling and regulating tissue-specific gene expression. Despite the rich literature on the importance of cooperative TFs for deciphering the mechanisms of individual regulatory programs that control tissue specificity in several organisms such as human, mouse, or Drosophila melanogaster, to date, there is still need for a comprehensive study to detect specific TF cooperations in regulatory processes of cattle tissues. To address the needs of knowledge about specific combinatorial gene regulation in cattle tissues, we made use of three publicly available RNA-seq datasets and obtained tissue-specific gene (TSG) sets for ten tissues (heart, lung, liver, kidney, duodenum, muscle tissue, adipose tissue, colon, spleen and testis). By analyzing these TSG-sets, tissue-specific TF cooperations of each tissue have been identified. The results reveal that similar to the combinatorial regulatory events of model organisms, TFs change their partners depending on their biological functions in different tissues. Particularly with regard to preferential partner choice of the transcription factors STAT3 and NR2C2, this phenomenon has been highlighted with their five different specific cooperation partners in multiple tissues. The information about cooperative TFs could be promising: i) to understand the molecular mechanisms of regulating processes; and ii) to extend the existing knowledge on the importance of single TFs in cattle tissues.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

