An algorithm for learning shape and appearance models without annotations
- PMID: 31096134
- PMCID: PMC6554617
- DOI: 10.1016/j.media.2019.04.008
An algorithm for learning shape and appearance models without annotations
Abstract
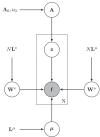
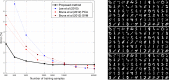
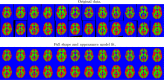
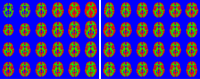
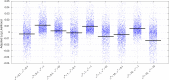
This paper presents a framework for automatically learning shape and appearance models for medical (and certain other) images. The algorithm was developed with the aim of eventually enabling distributed privacy-preserving analysis of brain image data, such that shared information (shape and appearance basis functions) may be passed across sites, whereas latent variables that encode individual images remain secure within each site. These latent variables are proposed as features for privacy-preserving data mining applications. The approach is demonstrated qualitatively on the KDEF dataset of 2D face images, showing that it can align images that traditionally require shape and appearance models trained using manually annotated data (manually defined landmarks etc.). It is applied to the MNIST dataset of handwritten digits to show its potential for machine learning applications, particularly when training data is limited. The model is able to handle "missing data", which allows it to be cross-validated according to how well it can predict left-out voxels. The suitability of the derived features for classifying individuals into patient groups was assessed by applying it to a dataset of over 1900 segmented T1-weighted MR images, which included images from the COBRE and ABIDE datasets.
Keywords: Appearance model; Diffeomorphisms; Geodesic shooting; Latent variables; Machine learning; Shape model.
Copyright © 2019 Wellcome Centre for Human Neuroimaging. Published by Elsevier B.V. All rights reserved.
Figures
















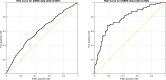
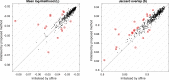

References
-
- Adams D., Rohlf F., Slice D. Geometric morphometrics: ten years of progress following the ‘revolution’. Ital. J. Zool. 2004;71(1):5–16.
- Adams, D., Rohlf, F., Slice, D., 2004. Geometric morphometrics: Ten years of progress following the ‘revolution’. Italian Journal of Zoology 71, 5–16.
-
- Allassonnière S., Amit Y., Trouve A. Towards a coherent statistical framework for dense deformable template estimation. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 2007;69(1):3–29.
- Allassonnière, S., Amit, Y., Trouve, A., 2007. Towards a coherent statistical framework for dense deformable template estimation. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 69, 3–29.
-
- Allassonnière S., Durrleman S., Kuhn E. Bayesian mixed effect atlas estimation with a diffeomorphic deformation model. SIAM J. Imaging Sci. 2015;8(3):1367–1395.
- Allassonnière, S., Durrleman, S., Kuhn, E., 2015. Bayesian mixed effect atlas estimation with a diffeomorphic deformation model. SIAM Journal on Imaging Sciences 8, 1367–1395.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

