Virion-Associated Cholesterol Regulates the Infection of Human Parainfluenza Virus Type 3
- PMID: 31096557
- PMCID: PMC6563303
- DOI: 10.3390/v11050438
Virion-Associated Cholesterol Regulates the Infection of Human Parainfluenza Virus Type 3
Abstract
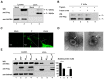
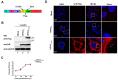
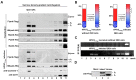
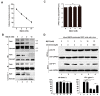
The matrix (M) proteins of paramyxoviruses bind to the nucleocapsids and cytoplasmic tails of glycoproteins, thus mediating the assembly and budding of virions. We first determined the budding characterization of the HPIV3 Fusion (F) protein to investigate the assembly mechanism of human parainfluenza virus type 3 (HPIV3). Our results show that expression of the HPIV3 F protein alone is sufficient to initiate the release of virus-like particles (VLPs), and the F protein can regulate the VLP-forming ability of the M protein. Furthermore, HPIV3F-Flag, which is a recombinant HPIV3 with a Flag tag at the C-terminus of the F protein, was constructed and recovered. We found that the M, F, and hemagglutinin-neuraminidase (HN) proteins and the viral genome can accumulate in lipid rafts in HPIV3F-Flag-infected cells, and the F protein mainly exists in the form of F1 in VLPs, lipid rafts, and purified virions. Furthermore, the function of cholesterol in the viral envelope and cell membrane was assessed via the elimination of cholesterol by methyl-β-cyclodextrin (MβCD). Our results suggest that the infectivity of HPIV3 was markedly reduced, due to defective internalization ability in the absence of cholesterol. These results reveal that HPIV3 might assemble in the lipid rafts to acquire cholesterol for the envelope of HPIV3, which suggests the that disruption of the cholesterol composition of HPIV3 virions might be a useful method for the design of anti-HPIV3 therapy.
Keywords: HPIV3; cholesterol; fusion protein; internalization; lipid rafts; viral assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
A leucine residue in the C terminus of human parainfluenza virus type 3 matrix protein is essential for efficient virus-like particle and virion release.J Virol. 2014 Nov;88(22):13173-88. doi: 10.1128/JVI.01485-14. Epub 2014 Sep 3. J Virol. 2014. PMID: 25187547 Free PMC article.
-
Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo.mBio. 2013 Oct 22;4(5):e00803-13. doi: 10.1128/mBio.00803-13. mBio. 2013. PMID: 24149514 Free PMC article.
-
Human parainfluenza virus 3 field strains undergo extracellular fusion protein cleavage to activate entry.mBio. 2024 Nov 13;15(11):e0232724. doi: 10.1128/mbio.02327-24. Epub 2024 Oct 9. mBio. 2024. PMID: 39382296 Free PMC article.
-
Interaction of Human Parainfluenza Virus Type 3 Nucleoprotein with Matrix Protein Mediates Internal Viral Protein Assembly.J Virol. 2015 Dec 9;90(5):2306-15. doi: 10.1128/JVI.02324-15. J Virol. 2015. PMID: 26656716 Free PMC article.
-
Adaptation of human parainfluenza virus to airway epithelium reveals fusion properties required for growth in host tissue.mBio. 2012 Jun 5;3(3):e00137-12. doi: 10.1128/mBio.00137-12. Print 2012. mBio. 2012. PMID: 22669629 Free PMC article.
Cited by
-
Lipid rafts disruption by statins negatively impacts the interaction between SARS-CoV-2 S1 subunit and ACE2 in intestinal epithelial cells.Front Microbiol. 2024 Jan 8;14:1335458. doi: 10.3389/fmicb.2023.1335458. eCollection 2023. Front Microbiol. 2024. PMID: 38260879 Free PMC article.
-
Polyamine-Linked Cholesterol Incorporation in Rift Valley Fever Virus Particles Promotes Infectivity.ACS Infect Dis. 2022 Aug 12;8(8):1439-1448. doi: 10.1021/acsinfecdis.2c00071. Epub 2022 Jul 5. ACS Infect Dis. 2022. PMID: 35786847 Free PMC article.
-
Cholesterol Modulates the Interaction between HIV-1 Viral Protein R and Membrane.Membranes (Basel). 2021 Oct 13;11(10):784. doi: 10.3390/membranes11100784. Membranes (Basel). 2021. PMID: 34677550 Free PMC article.
-
Natural small molecules as inhibitors of coronavirus lipid-dependent attachment to host cells: a possible strategy for reducing SARS-COV-2 infectivity?Acta Biomed. 2020 Mar 19;91(1):161-164. doi: 10.23750/abm.v91i1.9402. Acta Biomed. 2020. PMID: 32191676 Free PMC article.
-
Cholesterol, lipoproteins, and COVID-19: Basic concepts and clinical applications.Biochim Biophys Acta Mol Cell Biol Lipids. 2021 Feb;1866(2):158849. doi: 10.1016/j.bbalip.2020.158849. Epub 2020 Nov 4. Biochim Biophys Acta Mol Cell Biol Lipids. 2021. PMID: 33157278 Free PMC article. Review.
References
-
- Viard M., Parolini I., Sargiacomo M., Fecchi K., Ramoni C., Ablan S., Ruscetti F.W., Wang J.M., Blumenthal R. Role of cholesterol in human immunodeficiency virus type 1 envelope protein-mediated fusion with host cells. J. Virol. 2002;76:11584–11595. doi: 10.1128/JVI.76.22.11584-11595.2002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

