Identification of a regeneration-organizing cell in the Xenopus tail
- PMID: 31097661
- PMCID: PMC6986927
- DOI: 10.1126/science.aav9996
Identification of a regeneration-organizing cell in the Xenopus tail
Abstract
Unlike mammals, Xenopus laevis tadpoles have a high regenerative potential. To characterize this regenerative response, we performed single-cell RNA sequencing after tail amputation. By comparing naturally occurring regeneration-competent and -incompetent tadpoles, we identified a previously unrecognized cell type, which we term the regeneration-organizing cell (ROC). ROCs are present in the epidermis during normal tail development and specifically relocalize to the amputation plane of regeneration-competent tadpoles, forming the wound epidermis. Genetic ablation or manual removal of ROCs blocks regeneration, whereas transplantation of ROC-containing grafts induces ectopic outgrowths in early embryos. Transcriptional profiling revealed that ROCs secrete ligands associated with key regenerative pathways, signaling to progenitors to reconstitute lost tissue. These findings reveal the cellular mechanism through which ROCs form the wound epidermis and ensure successful regeneration.
Copyright © 2019 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
The authors declare no competing interests.
Figures


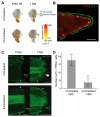


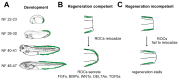
Comment in
-
Development: How Tadpoles ROC Tail Regeneration.Curr Biol. 2019 Aug 5;29(15):R756-R758. doi: 10.1016/j.cub.2019.06.026. Curr Biol. 2019. PMID: 31386855
References
-
- Tanaka EM. The Molecular and Cellular Choreography of Appendage Regeneration. Cell. 2016;165:1598–1608. - PubMed
-
- Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes & Development. 2007;21:1292–1315. - PubMed
-
- Beck CW, Christen B, Slack JMW. Molecular Pathways Needed for Regeneration of Spinal Cord and Muscle in a Vertebrate. Developmental Cell. 2003;5:429–439. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

