Molecular characterization of a Korean porcine epidemic diarrhea virus strain NB1
- PMID: 31097871
- PMCID: PMC6450166
Molecular characterization of a Korean porcine epidemic diarrhea virus strain NB1
Abstract
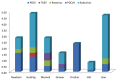
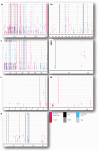
In Korea, for the past 30 years (1987-present), porcine epidemic diarrhea (PED) has been established as an endemic situation in which multiple genogroups of classical G1 and G2b, and the recently introduced pandemic G2a, coexisted. Because of the dynamic nature of the virus, continuous field monitoring for PEDV strains is required. This study is the first to reveal prevalence of PEDV in 9 sampling provinces, with an overall detection rate of 6.70%. Porcine endemic diarrhea virus (PEDV) was present in pigs of all ages, especially in the non-PED vaccinated groups. The highest detection rate was in the finisher group (2.34%), followed by that in the newborn group (1.56%). Secondly, using Sanger sequencing, this study recovered a complete genome (28 005 nucleotides long) of NB1 strain from a farm severely affected by PED. Analyses of nucleotide and deduced amino acid sequences showed that NB1 differed from 18 other Korean PEDV mostly in 4 protein coding genes: ORF1a, ORF1b, S, and N. Two amino acid substitutions (V635E and Y681Q) in the COE and S1D neutralizing epitopes of NB1 resulted in antigenic index alteration of the adjacent sites, one of which contributed to a mutation that escaped neutralizing antibodies.
En Corée, pour les 30 dernières années (1987 à ce jour), la diarrhée épidémique porcine (DEP) s’est établie comme une situation endémique dans laquelle de multiples génogroupes des classiques G1 et G2b, ainsi que le G2a pandémique récemment introduit, ont coexisté. Étant donné la nature dynamique du virus, un suivi continu sur le terrain des souches de DEP est requis. La présente étude est la première à révéler la prévalence de DEP dans neuf provinces échantillonnées, avec un taux de détection global de 6,70 %. Le virus de la DEP (VDEP) était présent chez les porcs de tout âge, spécialement dans les groupes d’animaux non-vaccinés contre la DEP. Les animaux dans le groupe en finition avaient taux de détection le plus élevé (2,34 %), suivi par ceux du groupe des nouveau-nés (1,56 %). Deuxièmement, en utilisant le séquençage de Sanger, nous avons récupéré un génome complet (28 005 nucléotides de long) de la souche NB1 sur une ferme sévèrement affectée par la DEP. L’analyse des nucléotides et des séquences d’acides aminés déduites a montré que NB1 différaient de 18 autres VDEP coréens principalement dans quatre gènes codant pour protéines: ORF1a, ORF1b, S, et N. Deux substitutions d’acides aminés (V635E et Y681Q) dans les épitopes neutralisants COE et S1D de NB1 ont résulté en une altération de l’index antigénique des sites adjacents, dont l’un contribuait à une mutation qui échappait aux anticorps neutralisants.(Traduit par Docteur Serge Messier).
Figures


References
-
- Duarte M, Gelfi J, Lambert P, Rasschaert D, Laude H. Genome organization of porcine epidemic diarrhoea virus. In: Laude H, Vautherot J-F, editors. Coronaviruses: Molecular Biology and Virus-Host Interactions. Boston, Massachusetts: Springer; 1993. pp. 55–60. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
