Two-Dimensional Separation Using High-pH and Low-pH Reversed Phase Liquid Chromatography for Top-down Proteomics
- PMID: 31097918
- PMCID: PMC6516780
- DOI: 10.1016/j.ijms.2017.09.001
Two-Dimensional Separation Using High-pH and Low-pH Reversed Phase Liquid Chromatography for Top-down Proteomics
Abstract
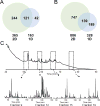
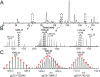
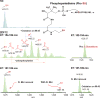
Advancements in chromatographic separation are critical to in-depth top-down proteomics of complex intact protein samples. Reversed-phase liquid chromatography is the most prevalent technique for top-down proteomics. However, in cases of high complexities and large dynamic ranges, 1D-RPLC may not provide sufficient coverage of the proteome. To address these challenges, orthogonal separation techniques are often combined to improve the coverage and the dynamic range of detection. In this study, a "salt-free" high-pH RPLC was evaluated as an orthogonal dimension of separation to conventional low-pH RPLC with top-down MS. The RPLC separations with low-pH conditions (pH=2) and high-pH conditions (pH=10) were compared to confirm the good orthogonality between high-pH and low-pH RPLC's. The offline 2D RPLC-RPLC-MS/MS analyses of intact E. coli samples were evaluated for the improvement of intact protein identifications as well as intact proteoform characterizations. Compared to the 163 proteins and 328 proteoforms identified using a 1D RPLC-MS approach, 365 proteins and 886 proteoforms were identified using the 2D RPLC-RPLC top-down MS approach. Our results demonstrate that the 2D RPLC-RPLC top-down approach holds great potential for in-depth top-down proteomics studies by utilizing the high resolving power of RPLC separations and by using mass spectrometry compatible buffers for easy sample handling for online MS analysis.
Keywords: 2D separation; Top-down proteomics; proteoform.
Figures
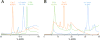


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
