Free-running 3D whole heart myocardial T1 mapping with isotropic spatial resolution
- PMID: 31099442
- PMCID: PMC6851769
- DOI: 10.1002/mrm.27811
Free-running 3D whole heart myocardial T1 mapping with isotropic spatial resolution
Abstract
Purpose: To develop a free-running (free-breathing, retrospective cardiac gating) 3D myocardial T1 mapping with isotropic spatial resolution.
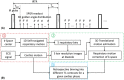
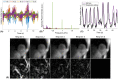
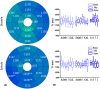
Methods: The free-running sequence is inversion recovery (IR)-prepared followed by continuous 3D golden angle radial data acquisition. 1D respiratory motion signal is extracted from the k-space center of all spokes and used to bin the k-space data into different respiratory states, enabling estimation and correction of 3D translational respiratory motion, whereas cardiac motion is recorded using electrocardiography and synchronized with data acquisition. 3D translational respiratory motion compensated T1 maps at diastole and systole were generated with 1.5 mm isotropic spatial resolution with low-rank inversion and high-dimensionality patch-based undersampled reconstruction. The technique was validated against conventional methods in phantom and 9 healthy subjects.
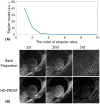
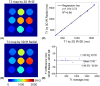
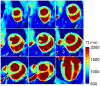
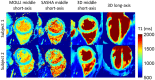
Results: Phantom results demonstrated good agreement (R2 = 0.99) of T1 estimation with reference method. Homogeneous systolic and diastolic 3D T1 maps were reconstructed from the proposed technique. Diastolic septal T1 estimated with the proposed method (1140 ± 36 ms) was comparable to the saturation recovery single-shot acquisition (SASHA) sequence (1153 ± 49 ms), but was higher than the modified Look-Locker inversion recovery (MOLLI) sequence (1037 ± 33 ms). Precision of the proposed method (42 ± 8 ms) was comparable to MOLLI (41 ± 7 ms) and improved with respect to SASHA (87 ± 19 ms).
Conclusions: The proposed free-running whole heart T1 mapping method allows for reconstruction of isotropic resolution 3D T1 maps at different cardiac phases, serving as a promising tool for whole heart myocardial tissue characterization.
Keywords: 3D radial; free-running; inversion recovery; myocardial T1 mapping.
© 2019 International Society for Magnetic Resonance in Medicine.
Figures


References
-
- Hamlin SA, Henry TS, Little BP, Lerakis S, Stillman AE. Mapping the future of cardiac MR imaging: case‐based review of T1 and T2 mapping techniques. Radiographics. 2014;34:1594–1611. - PubMed
-
- Schelbert EB, Messroghli DR. State of the art: clinical applications of cardiac T1 mapping. Radiology. 2016;278:658–676. - PubMed
-
- Messroghli DR, Moon JC, Ferreira VM, et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: a consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J Cardiovasc Magn Reson. 2017;19:75. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

