Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers
- PMID: 31100245
- PMCID: PMC6527355
- DOI: 10.1016/j.molcel.2019.04.025
Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers
Abstract
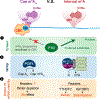
Cellular RNAs are naturally decorated with a variety of chemical modifications. The structural diversity of the modified nucleosides provides regulatory potential to sort groups of RNAs for organized metabolism and functions, thus affecting gene expression. Recent years have witnessed a burst of interest in and understanding of RNA modification biology, thanks to the emerging transcriptome-wide sequencing methods for mapping modified sites, highly sensitive mass spectrometry for precise modification detection and quantification, and extensive characterization of the modification "effectors," including enzymes ("writers" and "erasers") that alter the modification level and binding proteins ("readers") that recognize the chemical marks. However, challenges remain due to the vast heterogeneity in expression abundance of different RNA species, further complicated by divergent cell-type-specific and tissue-specific expression and localization of the effectors as well as modifications. In this review, we highlight recent progress in understanding the function of N6-methyladenosine (m6A), the most abundant internal mark on eukaryotic mRNA, in light of the specific biological contexts of m6A effectors. We emphasize the importance of context for RNA modification regulation and function.
Keywords: FTO; METTL14; METTL3; N(6)-methyladenosine; RNA modifications; YTHDF proteins; context-dependent functions; epitranscriptome; gene expression regulation; m(6)A.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures


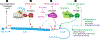
References
-
- Adams JM, and Cory S (1975). Modified nucleosides and bizarre 5[prime]-termini in mouse myeloma mRNA. Nature 255, 28–33. - PubMed
-
- Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, et al. (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

