A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface
- PMID: 31100268
- PMCID: PMC6629437
- DOI: 10.1016/j.cell.2019.04.011
A Site of Vulnerability on the Influenza Virus Hemagglutinin Head Domain Trimer Interface
Abstract
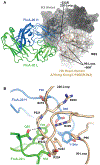
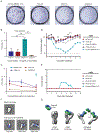
Here, we describe the discovery of a naturally occurring human antibody (Ab), FluA-20, that recognizes a new site of vulnerability on the hemagglutinin (HA) head domain and reacts with most influenza A viruses. Structural characterization of FluA-20 with H1 and H3 head domains revealed a novel epitope in the HA trimer interface, suggesting previously unrecognized dynamic features of the trimeric HA protein. The critical HA residues recognized by FluA-20 remain conserved across most subtypes of influenza A viruses, which explains the Ab's extraordinary breadth. The Ab rapidly disrupted the integrity of HA protein trimers, inhibited cell-to-cell spread of virus in culture, and protected mice against challenge with viruses of H1N1, H3N2, H5N1, or H7N9 subtypes when used as prophylaxis or therapy. The FluA-20 Ab has uncovered an exceedingly conserved protective determinant in the influenza HA head domain trimer interface that is an unexpected new target for anti-influenza therapeutics and vaccines.
Keywords: B-lymphocytes; antibodies; antibody-dependent cell cytotoxicity; antigen-antibody reactions; hemagglutinin glycoproteins; influenza A virus; influenza virus; monoclonal; viral.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures




Comment in
-
"Breathing" Hemagglutinin Reveals Cryptic Epitopes for Universal Influenza Vaccine Design.Cell. 2019 May 16;177(5):1086-1088. doi: 10.1016/j.cell.2019.04.034. Cell. 2019. PMID: 31100263 Free PMC article.
References
-
- Aiyegbo MS, Eli IM, Spiller BW, Williams DR, Kim R, Lee DE, Liu T, Li S, Stewart PL, and Crowe JE Jr. (2014). Differential accessibility of a rotavirus VP6 epitope in trimers comprising type I, II, or III channels as revealed by binding of a human rotavirus VP6-specific antibody. J. Virol 88, 469–476. - PMC - PubMed
-
- Al-Hubeshy ZB, Coleman A, Nelson M, and Goodier MR (2011). A rapid method for assessment of natural killer cell function after multiple receptor crosslinking. J. Immunol. Methods 366, 52–59. - PubMed
-
- Alter G, Malenfant JM, and Altfeld M (2004). CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods 294, 15–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

