Response to IL-6 trans- and IL-6 classic signalling is determined by the ratio of the IL-6 receptor α to gp130 expression: fusing experimental insights and dynamic modelling
- PMID: 31101051
- PMCID: PMC6525395
- DOI: 10.1186/s12964-019-0356-0
Response to IL-6 trans- and IL-6 classic signalling is determined by the ratio of the IL-6 receptor α to gp130 expression: fusing experimental insights and dynamic modelling
Abstract
Background: Interleukin-6 is a pleiotropic cytokine with high clinical relevance and an important mediator of cellular communication, orchestrating both pro- and anti-inflammatory processes. Interleukin-6-induced signalling is initiated by binding of IL-6 to the IL-6 receptor α and subsequent binding to the signal transducing receptor subunit gp130. This active receptor complex initiates signalling through the Janus kinase/signal transducer and activator of transcription pathway. Of note, IL-6 receptor α exists in a soluble and a transmembrane form. Binding of IL-6 to membrane-bound IL-6 receptor α induces anti-inflammatory classic signalling, whereas binding of IL-6 to soluble IL-6 receptor α induces pro-inflammatory trans-signalling. Trans-signalling has been described to be markedly stronger than classic signalling. Understanding the molecular mechanisms that drive differences between trans- and classic signalling is important for the design of trans-signalling-specific therapies. These differences will be addressed here using a combination of dynamic mathematical modelling and molecular biology.
Methods: We apply an iterative systems biology approach using set-based modelling and validation approaches combined with quantitative biochemical and cell biological analyses.
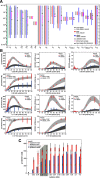
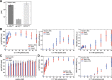
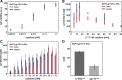
Results: The combination of experimental analyses and dynamic modelling allows to relate the observed differences between IL-6-induced trans- and classic signalling to cell-type specific differences in the expression and ratios of the individual subunits of the IL-6 receptor complex. Canonical intracellular Jak/STAT signalling is indifferent in IL-6-induced trans- and classic signalling.
Conclusion: This study contributes to the understanding of molecular mechanisms of IL-6 signal transduction and underlines the power of combined dynamical modelling, model-based validation and biological experiments. The opposing pro- and anti-inflammatory responses initiated by IL-6 trans- and classic signalling depend solely on the expression ratios of the subunits of the entire receptor complex. By pointing out the importance of the receptor expression ratio for the strength of IL-6 signalling this study lays a foundation for future precision medicine approaches that aim to selectively block pro-inflammatory trans-signalling. Furthermore, the derived models can be used for future therapy design.
Keywords: Classic signalling; Computational dynamic modelling; IL-6; IL-6 receptor α; Inflammation; Interleukin-6; Jak/STAT signalling; Set-based modelling and analysis; Signal transduction; Systems biology; Trans-signalling; gp130.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Andus T, Geiger T, Hirano T, Kishimoto T, Tran-Thi TA, Decker K, Heinrich PC. Regulation of synthesis and secretion of major rat acute-phase proteins by recombinant human interleukin-6 (BSF-2/IL-6) in hepatocyte primary cultures. Eur J Biochem. 1988;173:287–293. doi: 10.1111/j.1432-1033.1988.tb13997.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

