Identification of three subtypes of triple-negative breast cancer with potential therapeutic implications
- PMID: 31101122
- PMCID: PMC6525459
- DOI: 10.1186/s13058-019-1148-6
Identification of three subtypes of triple-negative breast cancer with potential therapeutic implications
Abstract
Background: Heterogeneity and lack of targeted therapies represent the two main impediments to precision treatment of triple-negative breast cancer (TNBC), and therefore, molecular subtyping and identification of therapeutic pathways are required to optimize medical care. The aim of the present study was to define robust TNBC subtypes with clinical relevance.
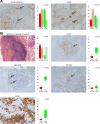
Methods: Gene expression profiling by means of DNA chips was conducted in an internal TNBC cohort composed of 238 patients. In addition, external data (n = 257), obtained by using the same DNA chip, were used for validation. Fuzzy clustering was followed by functional annotation of the clusters. Immunohistochemistry was used to confirm transcriptomics results: CD138 and CD20 were used to test for plasma cell and B lymphocyte infiltrations, respectively; MECA79 and CD31 for tertiary lymphoid structures; and UCHL1/PGP9.5 and S100 for neurogenesis.
Results: We identified three molecular clusters within TNBC: one molecular apocrine (C1) and two basal-like-enriched (C2 and C3). C2 presented pro-tumorigenic immune response (immune suppressive), high neurogenesis (nerve infiltration), and high biological aggressiveness. In contrast, C3 exhibited adaptive immune response associated with complete B cell differentiation that occurs in tertiary lymphoid structures, and immune checkpoint upregulation. External cohort subtyping by means of the same approach proved the robustness of these results. Furthermore, plasma cell and B lymphocyte infiltrates, tertiary lymphoid structures, and neurogenesis were validated at the protein levels by means of histological evaluation and immunohistochemistry.
Conclusion: Our work showed that TNBC can be subcategorized in three different subtypes characterized by marked biological features, some of which could be targeted by specific therapies.
Keywords: Breast cancer; Immunome; Molecular subtypes; Neurogenesis; Tertiary lymphoid structures; Transcriptomics; Triple-negative.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Bonsang-Kitzis H, Sadacca B, Hamy-Petit AS, Moarii M, Pinheiro A, Laurent C, et al. Biological network-driven gene selection identifies a stromal immune module as a key determinant of triple-negative breast carcinoma prognosis. Oncoimmunology. 2016;5:e1061176. doi: 10.1080/2162402X.2015.1061176. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

