A chemoprobe tracks its target
- PMID: 31101661
- PMCID: PMC6527164
- DOI: 10.1074/jbc.H119.008945
A chemoprobe tracks its target
Abstract
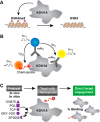
Small-molecule inhibitors of histone-modifying enzymes have significant clinical utility for managing diseases such as cancer. These inhibitors are usually identified and monitored through their effects on the gain or loss of specific histone marks. In cells, multiple related enzymes can place or remove a specific mark; therefore, relying on an indirect measure of inhibitor engagement can be misleading. Mascaró et al. describe a luminescence-based ELISA approach that directly monitors binding of inhibitors to the histone lysine demethylase KDM1A.
© 2019 Ansari.
Conflict of interest statement
The author declares that he has no conflicts of interest with the contents of this article
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

