An N-terminal-truncated isoform of FAM134B (FAM134B-2) regulates starvation-induced hepatic selective ER-phagy
- PMID: 31101736
- PMCID: PMC6526285
- DOI: 10.26508/lsa.201900340
An N-terminal-truncated isoform of FAM134B (FAM134B-2) regulates starvation-induced hepatic selective ER-phagy
Abstract
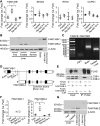
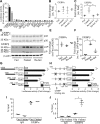
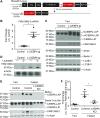
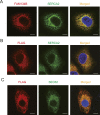
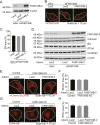
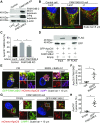
Autophagy is a conserved system that adapts to nutrient starvation, after which proteins and organelles are degraded to recycle amino acids in response to starvation. Recently, the ER was added to the list of targets of autophagic degradation. Autophagic degradation pathways of bulk ER and the specific proteins sorted through the ER are considered key mechanisms in maintaining ER homeostasis. Four ER-resident proteins (FAM134B, CCPG1, SEC62, and RTN3) have been identified as ER-resident cargo receptors, which contain LC3-interacting regions. In this study, we identified an N-terminal-truncated isoform of FAM134B (FAM134B-2) that contributes to starvation-induced ER-related autophagy. Hepatic FAM134B-2 but not full-length FAM134B (FAM134B-1) is expressed in a fed state. Starvation drastically induces FAM134B-2 but no other ER-resident cargo receptors through transcriptional activation by C/EBPβ. C/EBPβ overexpression increases FAM134B-2 recruitment into autophagosomes and lysosomal degradation. FAM134B-2 regulates lysosomal degradation of ER-retained secretory proteins such as ApoCIII. This study demonstrates that the C/EBPβ-FAM134B-2 axis regulates starvation-induced selective ER-phagy.
© 2019 Kohno et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
