Identification of Novel Susceptibility Loci and Genes for Prostate Cancer Risk: A Transcriptome-Wide Association Study in Over 140,000 European Descendants
- PMID: 31101764
- PMCID: PMC6606384
- DOI: 10.1158/0008-5472.CAN-18-3536
Identification of Novel Susceptibility Loci and Genes for Prostate Cancer Risk: A Transcriptome-Wide Association Study in Over 140,000 European Descendants
Abstract
Genome-wide association study-identified prostate cancer risk variants explain only a relatively small fraction of its familial relative risk, and the genes responsible for many of these identified associations remain unknown. To discover novel prostate cancer genetic loci and possible causal genes at previously identified risk loci, we performed a transcriptome-wide association study in 79,194 cases and 61,112 controls of European ancestry. Using data from the Genotype-Tissue Expression Project, we established genetic models to predict gene expression across the transcriptome for both prostate models and cross-tissue models and evaluated model performance using two independent datasets. We identified significant associations for 137 genes at P < 2.61 × 10-6, a Bonferroni-corrected threshold, including nine genes that remained significant at P < 2.61 × 10-6 after adjusting for all known prostate cancer risk variants in nearby regions. Of the 128 remaining associated genes, 94 have not yet been reported as potential target genes at known loci. We silenced 14 genes and many showed a consistent effect on viability and colony-forming efficiency in three cell lines. Our study provides substantial new information to advance our understanding of prostate cancer genetics and biology. SIGNIFICANCE: This study identifies novel prostate cancer genetic loci and possible causal genes, advancing our understanding of the molecular mechanisms that drive prostate cancer.
©2019 American Association for Cancer Research.
Figures
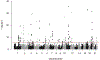
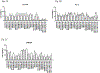
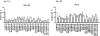
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians 2019;69:7–34 - PubMed
-
- Demichelis F, Stanford JL. Genetic predisposition to prostate cancer: Update and future perspectives. Urologic oncology 2015;33:75–84 - PubMed
-
- Crawford ED. Epidemiology of prostate cancer. Urology 2003;62:3–12 - PubMed

