Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism
- PMID: 31101807
- PMCID: PMC6525205
- DOI: 10.1038/s41467-019-09989-y
Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism
Abstract
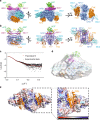
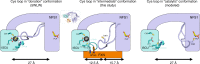
The core machinery for de novo biosynthesis of iron-sulfur clusters (ISC), located in the mitochondria matrix, is a five-protein complex containing the cysteine desulfurase NFS1 that is activated by frataxin (FXN), scaffold protein ISCU, accessory protein ISD11, and acyl-carrier protein ACP. Deficiency in FXN leads to the loss-of-function neurodegenerative disorder Friedreich's ataxia (FRDA). Here the 3.2 Å resolution cryo-electron microscopy structure of the FXN-bound active human complex, containing two copies of the NFS1-ISD11-ACP-ISCU-FXN hetero-pentamer, delineates the interactions of FXN with other component proteins of the complex. FXN binds at the interface of two NFS1 and one ISCU subunits, modifying the local environment of a bound zinc ion that would otherwise inhibit NFS1 activity in complexes without FXN. Our structure reveals how FXN facilitates ISC production through stabilizing key loop conformations of NFS1 and ISCU at the protein-protein interfaces, and suggests how FRDA clinical mutations affect complex formation and FXN activation.
Conflict of interest statement
Xiaodi Yu, Xidong Feng, Alain Martelli, Joseph F. Nabhan, Christine Bulawa, and Seungil Han are employees of Pfizer Inc. Nicolas G. Fox, Henry J. Bailey, Claire Strain-Damerell and Wyatt W. Yue declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

