Cellular consequences of arginine methylation
- PMID: 31101937
- PMCID: PMC6642692
- DOI: 10.1007/s00018-019-03140-2
Cellular consequences of arginine methylation
Abstract
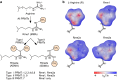
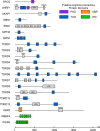
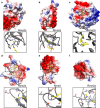
Arginine methylation is a ubiquitous post-translational modification. Three predominant types of arginine-guanidino methylation occur in Eukarya: mono (Rme1/MMA), symmetric (Rme2s/SDMA), and asymmetric (Rme2a/ADMA). Arginine methylation frequently occurs at sites of protein-protein and protein-nucleic acid interactions, providing specificity for binding partners and stabilization of important biological interactions in diverse cellular processes. Each methylarginine isoform-catalyzed by members of the protein arginine methyltransferase family, Type I (PRMT1-4,6,8) and Type II (PRMT5,9)-has unique downstream consequences. Methylarginines are found in ordered domains, domains of low complexity, and in intrinsically disordered regions of proteins-the latter two of which are intimately connected with biological liquid-liquid phase separation. This review highlights discoveries illuminating how arginine methylation affects genome integrity, gene transcription, mRNA splicing and mRNP biology, protein translation and stability, and phase separation. As more proteins and processes are found to be regulated by arginine methylation, its importance for understanding cellular physiology will continue to grow.
Keywords: Crosstalk; Histones; Liquid–liquid phase separation; Protein arginine methyltransferase; Ribonucleoprotein.
Figures

References
-
- Paik WK, Kim S. Protein methylase. I purification and properties of the enzyme. J Biol Chem. 1968;243(9):2108–2114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

