Structure-dependent activation of gene expression by bis-indole and quinoline-derived activators of nuclear receptor 4A2
- PMID: 31102570
- PMCID: PMC6791730
- DOI: 10.1111/cbdd.13564
Structure-dependent activation of gene expression by bis-indole and quinoline-derived activators of nuclear receptor 4A2
Abstract
Bis-indole derivatives including 1,1-bis(3'-indolyl)-1-(4-chlorophenyl)methane (DIM-C-pPhCl) and substituted quinolines such as chloroquine (CQ) and amodiaquine (AQ) are nuclear receptor 4A2 (NR4A2, Nurr1) ligands, and they exhibit anti-inflammatory activities in mouse and rat models of Parkinson's disease, respectively. However, computational modeling demonstrates that the quinoline derivatives interact with the ligand-binding domain, whereas the bis-indoles preferentially interact with a C-terminal cofactor binding site of NR4A2. In this study, the effects of DIM-C-pPhCl and related analogs were compared with CQ/AQ as inducers of NR4A2-responsive genes including vasoactive intestinal peptide, osteopontin, proopiomelanocortin, and neuropilin 1 in Panc1 and Panc28 pancreatic cancer cells. The results demonstrate that, among the bis-indole analogs, their relative potencies as inducers were structure-gene- and cell context dependent. In contrast, CQ and AQ were significantly less potent than the bis-indole derivatives and, for some of the NR4A2-regulated genes, CQ and AQ were inactive as inducers. These results demonstrate that although bis-indole and quinoline derivatives have been characterized as activators of NR4A2-dependent gene expression, these two classes of compounds exhibit different activities, indicating that they are selective NR4A2 modulators.
Keywords: NR4A2 ligand; bis-indole-derived; quinoline-derived.
© 2019 John Wiley & Sons A/S.
Conflict of interest statement
Figures
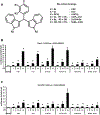

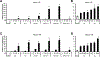
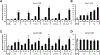
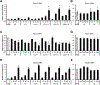
Similar articles
-
Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3'-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells.Biochem Pharmacol. 2012 May 15;83(10):1445-55. doi: 10.1016/j.bcp.2012.02.021. Epub 2012 Mar 3. Biochem Pharmacol. 2012. PMID: 22405837 Free PMC article.
-
Nuclear receptor 4A2 (NR4A2) is a druggable target for glioblastomas.J Neurooncol. 2020 Jan;146(1):25-39. doi: 10.1007/s11060-019-03349-y. Epub 2019 Nov 21. J Neurooncol. 2020. PMID: 31754919 Free PMC article.
-
Bis-Indole Derivatives as Dual Nuclear Receptor 4A1 (NR4A1) and NR4A2 Ligands.Biomolecules. 2024 Feb 27;14(3):284. doi: 10.3390/biom14030284. Biomolecules. 2024. PMID: 38540704 Free PMC article.
-
Potent synthetic and endogenous ligands for the adopted orphan nuclear receptor Nurr1.Exp Mol Med. 2021 Jan;53(1):19-29. doi: 10.1038/s12276-021-00555-5. Epub 2021 Jan 21. Exp Mol Med. 2021. PMID: 33479411 Free PMC article. Review.
-
Molecular Insights into NR4A2(Nurr1): an Emerging Target for Neuroprotective Therapy Against Neuroinflammation and Neuronal Cell Death.Mol Neurobiol. 2019 Aug;56(8):5799-5814. doi: 10.1007/s12035-019-1487-4. Epub 2019 Jan 25. Mol Neurobiol. 2019. PMID: 30684217 Review.
Cited by
-
Natural products and synthetic analogs as selective orphan nuclear receptor 4A (NR4A) modulators.Histol Histopathol. 2024 May;39(5):543-556. doi: 10.14670/HH-18-689. Epub 2023 Dec 13. Histol Histopathol. 2024. PMID: 38116863 Free PMC article. Review.
-
The Paradoxical Roles of Orphan Nuclear Receptor 4A (NR4A) in Cancer.Mol Cancer Res. 2021 Feb;19(2):180-191. doi: 10.1158/1541-7786.MCR-20-0707. Epub 2020 Oct 26. Mol Cancer Res. 2021. PMID: 33106376 Free PMC article. Review.
-
NR4A1 Ligands as Potent Inhibitors of Breast Cancer Cell and Tumor Growth.Cancers (Basel). 2021 May 29;13(11):2682. doi: 10.3390/cancers13112682. Cancers (Basel). 2021. PMID: 34072371 Free PMC article.
-
Analogs of the Dopamine Metabolite 5,6-Dihydroxyindole Bind Directly to and Activate the Nuclear Receptor Nurr1.ACS Chem Biol. 2021 Jul 16;16(7):1159-1163. doi: 10.1021/acschembio.1c00326. Epub 2021 Jun 24. ACS Chem Biol. 2021. PMID: 34165961 Free PMC article.
-
The Histone Methyltransferase Gene G9A Is Regulated by Nuclear Receptor 4A1 in Alveolar Rhabdomyosarcoma Cells.Mol Cancer Ther. 2021 Mar;20(3):612-622. doi: 10.1158/1535-7163.MCT-20-0474. Epub 2020 Dec 4. Mol Cancer Ther. 2021. PMID: 33277444 Free PMC article.
References
-
- De Miranda BR, Miller JA, Hansen RJ, Lunghofer PJ, Safe S, Gustafson DL, … Tjalkens RB. (2013). Neuroprotective efficacy and pharmacokinetic behavior of novel anti-inflammatory para-phenyl substituted diindolylmethanes in a mouse model of Parkinson’s disease. J Pharmacol Exp Ther, 345(1), 125–138. doi:10.1124/jpet.112.201558 - DOI - PMC - PubMed
-
- De Miranda BR, Popichak KA, Hammond SL, Jorgensen BA, Phillips AT, Safe S, & Tjalkens RB (2015a). The Nurr1 Activator 1,1-Bis(3’-Indolyl)-1-(p-Chlorophenyl)Methane Blocks Inflammatory Gene Expression in BV-2 Microglial Cells by Inhibiting Nuclear Factor kappaB. Mol Pharmacol, 87(6), 1021–1034. doi:10.1124/mol.114.095398 - DOI - PMC - PubMed
-
- De Miranda BR, Popichak KA, Hammond SL, Miller JA, Safe S, & Tjalkens RB (2015b). Novel para-phenyl substituted diindolylmethanes protect against MPTP neurotoxicity and suppress glial activation in a mouse model of Parkinson’s disease. Toxicol Sci, 143(2), 360–373. doi:10.1093/toxsci/kfu236 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

