Fingerprinting Orientation Distribution Functions in diffusion MRI detects smaller crossing angles
- PMID: 31102735
- PMCID: PMC7336899
- DOI: 10.1016/j.neuroimage.2019.05.024
Fingerprinting Orientation Distribution Functions in diffusion MRI detects smaller crossing angles
Abstract
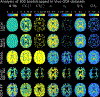
Diffusion tractography is routinely used to study white matter architecture and brain connectivity in vivo. A key step for successful tractography of neuronal tracts is the correct identification of tract directions in each voxel. Here we propose a fingerprinting-based methodology to identify these fiber directions in Orientation Distribution Functions, dubbed ODF-Fingerprinting (ODF-FP). In ODF-FP, fiber configurations are selected based on the similarity between measured ODFs and elements in a pre-computed library. In noisy ODFs, the library matching algorithm penalizes the more complex fiber configurations. ODF simulations and analysis of bootstrapped partial and whole-brain in vivo datasets show that the ODF-FP approach improves the detection of fiber pairs with small crossing angles while maintaining fiber direction precision, which leads to better tractography results. Rather than focusing on the ODF maxima, the ODF-FP approach uses the whole ODF shape to infer fiber directions to improve the detection of fiber bundles with small crossing angle. The resulting fiber directions aid tractography algorithms in accurately displaying neuronal tracts and calculating brain connectivity.
Keywords: Crossing angle; Diffusion MRI; Fiber identification; Fiber tractography; Fingerprinting; Multi-shell Q-ball imaging; Orientation distribution function; Radial diffusion spectrum imaging.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures


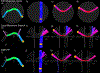

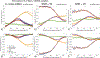





References
-
- Aja-Fernandez S, Alberola-Lopez C, Westin CF, 2008. Noise and signal estimation in magnitude mri and rician distributed images: a lmmse approach. IEEE Trans Image Process 17 (8), 1383–98. - PubMed
-
- Alexander D, Hubbard P, Hall M, Moore E, Ptito M, Parker G, Dyrby T, 2010. Orientationally invariant indices of axon diameter and density from diffusion mri. NeuroImage 52, 1374–89. - PubMed
-
- Alexander D, Pierpaoli C, Basser PJ, Gee J, 2001. Spatial transformations ofdiffusion tensor magnetic resonance images. IEEE Trans Med Imaging 20 (11), 1131–39. - PubMed
-
- Assaf Y, Basser P, 2005. Composite hindered and restricted model of diffusion (charmed) mr imaging of the human brain. NeuroImage 27, 48–58. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

