Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation
- PMID: 31103420
- PMCID: PMC7611809
- DOI: 10.1016/j.molcel.2019.04.029
Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation
Abstract
Regulatory sequences or erroneous incorporations during DNA transcription cause RNA polymerase backtracking and inactivation in all kingdoms of life. Reactivation requires RNA transcript cleavage. Essential transcription factors (GreA and GreB, or TFIIS) accelerate this reaction. We report four cryo-EM reconstructions of Escherichia coli RNA polymerase representing the entire reaction pathway: (1) a backtracked complex; a backtracked complex with GreB (2) before and (3) after RNA cleavage; and (4) a reactivated, substrate-bound complex with GreB before RNA extension. Compared with eukaryotes, the backtracked RNA adopts a different conformation. RNA polymerase conformational changes cause distinct GreB states: a fully engaged GreB before cleavage; a disengaged GreB after cleavage; and a dislodged, loosely bound GreB removed from the active site to allow RNA extension. These reconstructions provide insight into the catalytic mechanism and dynamics of RNA cleavage and extension and suggest how GreB targets backtracked complexes without interfering with canonical transcription.
Keywords: GreB; RNA polymerase structure; backtracking; cryo-EM; transcription; transcriptional arrest; transcriptional rescue.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


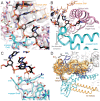
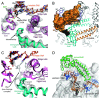

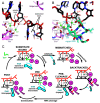
References
-
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–112. - PubMed
-
- Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–12355. - PubMed
-
- Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E coli. Cell. 1993;72:459–466. - PubMed
-
- Cheung ACM, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:–253 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

