Derivation, Validation, and Potential Treatment Implications of Novel Clinical Phenotypes for Sepsis
- PMID: 31104070
- PMCID: PMC6537818
- DOI: 10.1001/jama.2019.5791
Derivation, Validation, and Potential Treatment Implications of Novel Clinical Phenotypes for Sepsis
Abstract
Importance: Sepsis is a heterogeneous syndrome. Identification of distinct clinical phenotypes may allow more precise therapy and improve care.
Objective: To derive sepsis phenotypes from clinical data, determine their reproducibility and correlation with host-response biomarkers and clinical outcomes, and assess the potential causal relationship with results from randomized clinical trials (RCTs).
Design, settings, and participants: Retrospective analysis of data sets using statistical, machine learning, and simulation tools. Phenotypes were derived among 20 189 total patients (16 552 unique patients) who met Sepsis-3 criteria within 6 hours of hospital presentation at 12 Pennsylvania hospitals (2010-2012) using consensus k means clustering applied to 29 variables. Reproducibility and correlation with biological parameters and clinical outcomes were assessed in a second database (2013-2014; n = 43 086 total patients and n = 31 160 unique patients), in a prospective cohort study of sepsis due to pneumonia (n = 583), and in 3 sepsis RCTs (n = 4737).
Exposures: All clinical and laboratory variables in the electronic health record.
Main outcomes and measures: Derived phenotype (α, β, γ, and δ) frequency, host-response biomarkers, 28-day and 365-day mortality, and RCT simulation outputs.
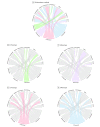
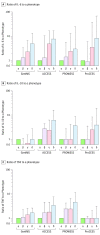
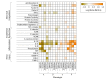
Results: The derivation cohort included 20 189 patients with sepsis (mean age, 64 [SD, 17] years; 10 022 [50%] male; mean maximum 24-hour Sequential Organ Failure Assessment [SOFA] score, 3.9 [SD, 2.4]). The validation cohort included 43 086 patients (mean age, 67 [SD, 17] years; 21 993 [51%] male; mean maximum 24-hour SOFA score, 3.6 [SD, 2.0]). Of the 4 derived phenotypes, the α phenotype was the most common (n = 6625; 33%) and included patients with the lowest administration of a vasopressor; in the β phenotype (n = 5512; 27%), patients were older and had more chronic illness and renal dysfunction; in the γ phenotype (n = 5385; 27%), patients had more inflammation and pulmonary dysfunction; and in the δ phenotype (n = 2667; 13%), patients had more liver dysfunction and septic shock. Phenotype distributions were similar in the validation cohort. There were consistent differences in biomarker patterns by phenotype. In the derivation cohort, cumulative 28-day mortality was 287 deaths of 5691 unique patients (5%) for the α phenotype; 561 of 4420 (13%) for the β phenotype; 1031 of 4318 (24%) for the γ phenotype; and 897 of 2223 (40%) for the δ phenotype. Across all cohorts and trials, 28-day and 365-day mortality were highest among the δ phenotype vs the other 3 phenotypes (P < .001). In simulation models, the proportion of RCTs reporting benefit, harm, or no effect changed considerably (eg, varying the phenotype frequencies within an RCT of early goal-directed therapy changed the results from >33% chance of benefit to >60% chance of harm).
Conclusions and relevance: In this retrospective analysis of data sets from patients with sepsis, 4 clinical phenotypes were identified that correlated with host-response patterns and clinical outcomes, and simulations suggested these phenotypes may help in understanding heterogeneity of treatment effects. Further research is needed to determine the utility of these phenotypes in clinical care and for informing trial design and interpretation.
Conflict of interest statement
Figures



Comment in
-
New Phenotypes for Sepsis: The Promise and Problem of Applying Machine Learning and Artificial Intelligence in Clinical Research.JAMA. 2019 May 28;321(20):1981-1982. doi: 10.1001/jama.2019.5794. JAMA. 2019. PMID: 31104067 No abstract available.
-
Identifying Sepsis Phenotypes.JAMA. 2019 Oct 8;322(14):1416. doi: 10.1001/jama.2019.12587. JAMA. 2019. PMID: 31593265 No abstract available.
-
Identifying Sepsis Phenotypes.JAMA. 2019 Oct 8;322(14):1416-1417. doi: 10.1001/jama.2019.12591. JAMA. 2019. PMID: 31593266 No abstract available.
-
Patient selection in sepsis: precision medicine using phenotypes and its implications for future clinical trial design.J Thorac Dis. 2019 Sep;11(9):3672-3675. doi: 10.21037/jtd.2019.09.31. J Thorac Dis. 2019. PMID: 31656636 Free PMC article. No abstract available.
-
Host-targeted approaches to sepsis due to community-acquired pneumonia.EBioMedicine. 2022 Dec;86:104335. doi: 10.1016/j.ebiom.2022.104335. Epub 2022 Dec 2. EBioMedicine. 2022. PMID: 36470827 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

