BIRC5 Gene Disruption via CRISPR/Cas9n Platform Suppress Acute Myelocytic Leukemia Progression
- PMID: 31104397
- PMCID: PMC6800533
- DOI: 10.29252/ibj.23.6.369
BIRC5 Gene Disruption via CRISPR/Cas9n Platform Suppress Acute Myelocytic Leukemia Progression
Abstract
Background: Acute myelocytic leukemia (AML) is a clonal malignancy resulting from the accumulation of genetic abnormalities in the cells. Human baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5), encodes survivin, is one of only a handful of genes that is differentially over-expressed in numerous malignant diseases including AML.
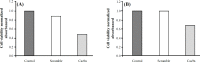
Methods: The BIRC5 was silenced permanently in two AML cell lines, HL‑60 and KG-1, via the CRISPR/Cas9n system. After transfection of CRISPR constructs, genomic DNA was extracted and amplified to assess mutation detection. To evaluate BIRC5 gene expression, quantitative real-time PCR was performed. Also, MTT cell viability and Annexin‑V/propidium iodide flowcytometric staining were performed, and the data were analyzed using the Kolmogorov-Smirnov, Levene's, and ANOVA tests.
Results: The results indicated that Cas9n and its sgRNAs successfully triggered site-specific cleavage and mutation in the BIRC5 gene locus. Moreover, suppression of BIRC5 resulted in the reduction of cell viability, and induction of apoptosis and necrosis in HL60 and KG1 suggested that the permanent suppression of BIRC5 remarkably dropped the gene expression and cells viability.
Conclusion: This study reinforces the idea that BIRC5 disruption via Cas9n:sgRNAs has favorable effects on the AML clinical outcome. It thereby can be a promising candidate in a variety of leukemia treatments.
Keywords: Acute myelocytic leukemia; CRISPR; Gene editing; Survivin.
Figures




Similar articles
-
Knockout Of BIRC5 Gene By CRISPR/Cas9 Induces Apoptosis And Inhibits Cell Proliferation In Leukemic Cell Lines, HL60 And KG1.Blood Lymphat Cancer. 2019 Nov 27;9:53-61. doi: 10.2147/BLCTT.S230383. eCollection 2019. Blood Lymphat Cancer. 2019. PMID: 31819702 Free PMC article.
-
Survivin' Acute Myeloid Leukaemia-A Personalised Target for inv(16) Patients.Int J Mol Sci. 2021 Sep 28;22(19):10482. doi: 10.3390/ijms221910482. Int J Mol Sci. 2021. PMID: 34638823 Free PMC article.
-
BIRC5 (survivin) splice variant expression correlates with refractory disease and poor outcome in pediatric acute myeloid leukemia: a report from the Children's Oncology Group.Pediatr Blood Cancer. 2014 Apr;61(4):647-52. doi: 10.1002/pbc.24822. Epub 2013 Oct 11. Pediatr Blood Cancer. 2014. PMID: 24127439 Free PMC article.
-
sgRNA Sequence Motifs Blocking Efficient CRISPR/Cas9-Mediated Gene Editing.Cell Rep. 2019 Jan 29;26(5):1098-1103.e3. doi: 10.1016/j.celrep.2019.01.024. Cell Rep. 2019. PMID: 30699341 Free PMC article.
-
Disruption of CTCF Boundary at HOXA Locus Promote BET Inhibitors' Therapeutic Sensitivity in Acute Myeloid Leukemia.Stem Cell Rev Rep. 2020 Dec;16(6):1280-1291. doi: 10.1007/s12015-020-10057-y. Epub 2020 Oct 14. Stem Cell Rev Rep. 2020. PMID: 33057942
Cited by
-
Ocoxin Increases the Antitumor Effect of BRAF Inhibition and Reduces Cancer Associated Fibroblast-Mediated Chemoresistance and Protumoral Activity in Metastatic Melanoma.Nutrients. 2021 Feb 21;13(2):686. doi: 10.3390/nu13020686. Nutrients. 2021. PMID: 33669949 Free PMC article.
-
Knockout Of BIRC5 Gene By CRISPR/Cas9 Induces Apoptosis And Inhibits Cell Proliferation In Leukemic Cell Lines, HL60 And KG1.Blood Lymphat Cancer. 2019 Nov 27;9:53-61. doi: 10.2147/BLCTT.S230383. eCollection 2019. Blood Lymphat Cancer. 2019. PMID: 31819702 Free PMC article.
-
Visualization Analysis of CRISPR Gene-editing Knowledge Map based on Citespace.Biol Bull Russ Acad Sci. 2021;48(6):705-720. doi: 10.1134/S1062359021060108. Epub 2021 Dec 17. Biol Bull Russ Acad Sci. 2021. PMID: 34955625 Free PMC article.
-
CRISPR-Cas9 in basic and translational aspects of cancer therapy.Bioimpacts. 2024;14(6):30087. doi: 10.34172/bi.2024.30087. Epub 2024 Mar 10. Bioimpacts. 2024. PMID: 39493894 Free PMC article. Review.
-
Differentially expressed autophagy-related genes are potential prognostic and diagnostic biomarkers in clear-cell renal cell carcinoma.Aging (Albany NY). 2019 Oct 17;11(20):9025-9042. doi: 10.18632/aging.102368. Epub 2019 Oct 17. Aging (Albany NY). 2019. PMID: 31626592 Free PMC article.
References
-
- Laosai J, Kosin C. Classification of acute leukemia using CD markers. presented at 8th International Conference on Knowledge and Smart Technology (KST), Chiangmai, 2016; Thailand: IEEE;
-
- El-Shafey AS, Amer SM, Allam NG, EL-Alfy MS. Hematological studies in Egyptian children with acute lymphoid and myeloid leukemia. International journal of advanced biomedicine. 2017;2(1):19–29.
-
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, Gundem G, Van Loo P, Martincorena I, Ganly P, Mudie L, McLaren S, O'Meara S, Raine K, Jones DR, Teague JW, Butler AP, Greaves MF, Ganser A, Döhner K, Schlenk RF, Döhner H, Campbell PJ. Genomic classification and prognosis in acute myeloid leukemia. The new England journal of medicine. 2016;374:2209–2221. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
