Quantitative Interactome Proteomics Reveals a Molecular Basis for ATF6-Dependent Regulation of a Destabilized Amyloidogenic Protein
- PMID: 31105062
- PMCID: PMC6642020
- DOI: 10.1016/j.chembiol.2019.04.001
Quantitative Interactome Proteomics Reveals a Molecular Basis for ATF6-Dependent Regulation of a Destabilized Amyloidogenic Protein
Abstract
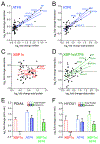
Activation of the unfolded protein response (UPR)-associated transcription factor ATF6 has emerged as a promising strategy to reduce the secretion and subsequent toxic aggregation of destabilized, amyloidogenic proteins implicated in systemic amyloid diseases. However, the molecular mechanism by which ATF6 activation reduces the secretion of amyloidogenic proteins remains poorly defined. We employ a quantitative interactomics platform to define how ATF6 activation reduces secretion of a destabilized, amyloidogenic immunoglobulin light chain (LC) associated with light-chain amyloidosis (AL). Using this platform, we show that ATF6 activation increases the targeting of this destabilized LC to a subset of pro-folding ER proteostasis factors that retains the amyloidogenic LC within the ER, preventing its secretion. Our results define a molecular basis for the ATF6-dependent reduction in destabilized LC secretion and highlight the advantage for targeting this UPR-associated transcription factor to reduce secretion of destabilized, amyloidogenic proteins implicated in AL and related systemic amyloid diseases.
Keywords: ATF6; ER chaperones; ER proteostasis; ER quality control; Unfolded Protein Response (UPR); XBP1s; quantitative proteomics.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
DECLARATION OF INTEREST
The authors declare that they have no competing interest.
Figures




Comment in
-
Interactome Changes Quantified to Identify the ER Proteostasis Network to Fight Amyloid Diseases.Cell Chem Biol. 2019 Jul 18;26(7):909-910. doi: 10.1016/j.chembiol.2019.07.003. Cell Chem Biol. 2019. PMID: 31323219 Free PMC article.
References
-
- Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, and Mori K (2008). ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct 33, 75–89. - PubMed
-
- Arendt BK, Ramirez-Alvarado M, Sikkink LA, Keats JJ, Ahmann GJ, Dispenzieri A, Fonseca R, Ketterling RP, Knudson RA, Mulvihill EM, et al. (2008). Biologic and genetic characterization of the novel amyloidogenic lambda light chain-secreting human cell lines, ALMC-1 and ALMC-2. Blood 112, 1931–1941. - PMC - PubMed
-
- Balchin D, Hayer-Hartl M, and Hartl FU (2016). In vivo aspects of protein folding and quality control. Science 353, aac4354. - PubMed

