Directed Evolution and Engineering of Gallium-Binding Phage Clones-A Preliminary Study
- PMID: 31105220
- PMCID: PMC6630928
- DOI: 10.3390/biomimetics4020035
Directed Evolution and Engineering of Gallium-Binding Phage Clones-A Preliminary Study
Abstract
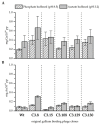
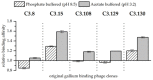
The phage surface display technology is a useful tool to screen and to extend the spectrum of metal-binding protein structures provided by nature. The directed evolution approach allows identifying specific peptide ligands for metals that are less abundant in the biosphere. Such peptides are attractive molecules in resource technology. For example, gallium-binding peptides could be applied to recover gallium from low concentrated industrial wastewater. In this study, we investigated the affinity and selectivity of five bacteriophage clones displaying different gallium-binding peptides towards gallium and arsenic in independent biosorption experiments. The displayed peptides were highly selective towards Ga3+ whereby long linear peptides showed a lower affinity and specificity than those with a more rigid structure. Cysteine scanning was performed to determine the relationship between secondary peptide structure and gallium sorption. By site-directed mutagenesis, the amino acids of a preselected peptide sequence are systematically replaced by cysteines. The resulting disulphide bridge considerably reduces the flexibility of linear peptides. Subsequent biosorption experiments carried out with the mutants obtained from cysteine scanning demonstrated, depending on the position of the cysteines in the peptide, either a considerable increase in the affinity of gallium compared to arsenic or an increase in the affinity for arsenic compared to gallium. This study shows the impressive effect on peptide-target interaction based on peptide structure and amino acid position and composition via the newly established systematic cysteine scanning approach.
Keywords: cysteine; gallium; metal–peptide interaction; peptide structure; phage surface display; site-directed mutagenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Chromatopanning for the identification of gallium binding peptides.J Chromatogr A. 2019 Aug 30;1600:158-166. doi: 10.1016/j.chroma.2019.04.037. Epub 2019 Apr 15. J Chromatogr A. 2019. PMID: 31040030
-
Gallium-binding peptides as a tool for the sustainable treatment of industrial waste streams.J Hazard Mater. 2021 Jul 15;414:125366. doi: 10.1016/j.jhazmat.2021.125366. Epub 2021 Feb 17. J Hazard Mater. 2021. PMID: 33636447
-
Biopanning of polypeptides binding to bovine ephemeral fever virus G1 protein from phage display peptide library.BMC Vet Res. 2018 Jan 4;14(1):3. doi: 10.1186/s12917-017-1315-x. BMC Vet Res. 2018. PMID: 29301517 Free PMC article.
-
M13 bacteriophage displaying disulfide-constrained microproteins.Gene. 1993 Jun 15;128(1):29-36. doi: 10.1016/0378-1119(93)90149-w. Gene. 1993. PMID: 8508957 Review.
-
Phage display: concept, innovations, applications and future.Biotechnol Adv. 2010 Nov-Dec;28(6):849-58. doi: 10.1016/j.biotechadv.2010.07.004. Epub 2010 Jul 23. Biotechnol Adv. 2010. PMID: 20659548 Review.
Cited by
-
Synthetic bacteria designed using ars operons: a promising solution for arsenic biosensing and bioremediation.World J Microbiol Biotechnol. 2024 May 6;40(6):192. doi: 10.1007/s11274-024-04001-2. World J Microbiol Biotechnol. 2024. PMID: 38709285 Review.
-
Application of Next Generation Sequencing (NGS) in Phage Displayed Peptide Selection to Support the Identification of Arsenic-Binding Motifs.Viruses. 2020 Nov 27;12(12):1360. doi: 10.3390/v12121360. Viruses. 2020. PMID: 33261041 Free PMC article.
References
-
- Farrell N. Biomedical uses and applications of inorganic chemistry. An overview. Coord. Chem. Rev. 2002;232:1–4. doi: 10.1016/S0010-8545(02)00100-5. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

