Envelope-Specific Recognition Patterns of HIV Vaccine-Induced IgG Antibodies Are Linked to Immunogen Structure and Sequence
- PMID: 31105688
- PMCID: PMC6492543
- DOI: 10.3389/fimmu.2019.00717
Envelope-Specific Recognition Patterns of HIV Vaccine-Induced IgG Antibodies Are Linked to Immunogen Structure and Sequence
Abstract
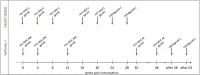
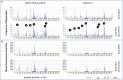
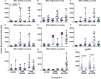
Background: A better understanding of the parameters influencing vaccine-induced IgG recognition of individual antigenic regions and their variants within the HIV Envelope protein (Env) can help to improve design of preventive HIV vaccines. Methods: Env-specific IgG responses were mapped in samples of the UKHVC003 Standard Group (UK003SG, n = 11 from UK) and TaMoVac01 (TMV01, n = 17 from Tanzania) HIV vaccine trials. Both trials consisted of three immunizations with DNA, followed by two boosts with recombinant Modified Vaccinia Virus Ankara (MVA), either mediating secretion of gp120 (UK003SG) or the presentation of cell membrane bound gp150 envelopes (TMV01) from infected cells, and an additional two boosts with 5 μg of CN54gp140 protein adjuvanted with glucopyranosyl lipid adjuvant (GLA). Env immunogen sequences in UK003SG were solely based on the clade C isolate CN54, whereas in TMV01 these were based on clades A, C, B, and CRF01AE. The peptide microarray included 8 globally representative Env sequences, CN54gp140 and the MVA-encoded Env immunogens from both trials, as well as additional peptide variants for hot spots of immune recognition. Results: After the second MVA boost, UK003SG vaccinees almost exclusively targeted linear, non-glycosylated antigenic regions located in the inter-gp120 interface. In contrast, TMV01 recipients most strongly targeted the V2 region and an immunodominant region in gp41. The V3 region was frequently targeted in both trials, with a higher recognition magnitude for diverse antigenic variants observed in the UK003SG (p < 0.0001). After boosting with CN54gp140/GLA, the overall response magnitude increased with a more comparable recognition pattern of antigenic regions and variants between the two trials. Recognition of most immunodominant regions within gp120 remained significantly stronger in UK003SG, whereas V2-region recognition was not boosted in either group. Conclusions: IgG recognition of linear antigenic Env regions differed between the two trials particularly after the second MVA boost. Structural features of the MVA-encoded immunogens, such as secreted, monomeric gp120 vs. membrane-anchored, functional gp150, and differences in prime-boost immunogen sequence variability most probably contributed to these differences. Prime-boosting with multivalent Env immunogens during TMV01 did not improve variant cross-recognition of immunodominant peptide variants in the V3 region.
Keywords: HIV; envelope-specific antibodies; epitope variant recognition; immunogen sequence; immunogen structure; vaccine.
Figures


Similar articles
-
Induction of Identical IgG HIV-1 Envelope Epitope Recognition Patterns After Initial HIVIS-DNA/MVA-CMDR Immunization and a Late MVA-CMDR Boost.Front Immunol. 2020 Apr 28;11:719. doi: 10.3389/fimmu.2020.00719. eCollection 2020. Front Immunol. 2020. PMID: 32411138 Free PMC article. Clinical Trial.
-
Boosting with Subtype C CN54rgp140 Protein Adjuvanted with Glucopyranosyl Lipid Adjuvant after Priming with HIV-DNA and HIV-MVA Is Safe and Enhances Immune Responses: A Phase I Trial.PLoS One. 2016 May 18;11(5):e0155702. doi: 10.1371/journal.pone.0155702. eCollection 2016. PLoS One. 2016. PMID: 27192151 Free PMC article. Clinical Trial.
-
A Trimeric HIV-1 Envelope gp120 Immunogen Induces Potent and Broad Anti-V1V2 Loop Antibodies against HIV-1 in Rabbits and Rhesus Macaques.J Virol. 2018 Feb 12;92(5):e01796-17. doi: 10.1128/JVI.01796-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237847 Free PMC article.
-
Structure-based vaccine design in HIV: blind men and the elephant?Curr Pharm Des. 2010;16(33):3744-53. doi: 10.2174/138161210794079173. Curr Pharm Des. 2010. PMID: 21128885 Free PMC article. Review.
-
Back to the future: covalent epitope-based HIV vaccine development.Expert Rev Vaccines. 2010 Sep;9(9):1027-43. doi: 10.1586/erv.10.77. Expert Rev Vaccines. 2010. PMID: 20822346 Free PMC article. Review.
Cited by
-
Induction of Identical IgG HIV-1 Envelope Epitope Recognition Patterns After Initial HIVIS-DNA/MVA-CMDR Immunization and a Late MVA-CMDR Boost.Front Immunol. 2020 Apr 28;11:719. doi: 10.3389/fimmu.2020.00719. eCollection 2020. Front Immunol. 2020. PMID: 32411138 Free PMC article. Clinical Trial.
-
Systematic comparison of HIV-1 Envelope-specific IgG responses induced by different vaccination regimens: Can we steer IgG recognition towards regions of viral vulnerability?Front Immunol. 2023 Jan 9;13:1075606. doi: 10.3389/fimmu.2022.1075606. eCollection 2022. Front Immunol. 2023. PMID: 36741409 Free PMC article.
-
Applications of Peptide Microarrays in Autoantibody, Infection, and Cancer Detection.Methods Mol Biol. 2023;2578:1-15. doi: 10.1007/978-1-0716-2732-7_1. Methods Mol Biol. 2023. PMID: 36152276 Review.
-
Rapid Response to Pandemic Threats: Immunogenic Epitope Detection of Pandemic Pathogens for Diagnostics and Vaccine Development Using Peptide Microarrays.J Proteome Res. 2020 Nov 6;19(11):4339-4354. doi: 10.1021/acs.jproteome.0c00484. Epub 2020 Sep 21. J Proteome Res. 2020. PMID: 32892628 Free PMC article. Review.
-
Stepwise Conformational Stabilization of a HIV-1 Clade C Consensus Envelope Trimer Immunogen Impacts the Profile of Vaccine-Induced Antibody Responses.Vaccines (Basel). 2021 Jul 6;9(7):750. doi: 10.3390/vaccines9070750. Vaccines (Basel). 2021. PMID: 34358165 Free PMC article.
References
-
- Gottardo R, Bailer RT, Korber BT, Gnanakaran S, Phillips J, Shen X, et al. . Plasma IgG to linear epitopes in the V2 and V3 regions of HIV-1 gp120 correlate with a reduced risk of infection in the RV144 vaccine efficacy trial. PLoS ONE. (2013) 8:e75665. 10.1371/journal.pone.0075665 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

