Developmental features of DNA methylation in CpG islands of human gametes and preimplantation embryos
- PMID: 31105782
- PMCID: PMC6507515
- DOI: 10.3892/etm.2019.7523
Developmental features of DNA methylation in CpG islands of human gametes and preimplantation embryos
Abstract
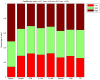
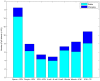
The aim of current study was to apply the methylated DNA immunoprecipitation (MeDIP)-Chip method to investigate dynamic changes in CpG island methylation in human sperm, oocytes and various developmental stages of preimplantation embryos. Samples were divided into eight groups: 1, sperm (n=30); 2, MII oocyte (n=25); 3, two-pronuclear (2PN) period zygote (n=25); 4, 4-cell stage embryo (n=5); 5, 8-cell stage embryo (n=4); 6, morula embryo (n=6); 7, blastular inner cell mass (ICM) group (n=5); 8, blastular trophoblastic cells (TE) (n=5). DNA was extracted and hybridized to NimbleGen Human DNA microarray. Following this, chip methylation data were read and analyzed. The CpG island methylation level of sperm was highest (peak value=15604), followed by oocytes (peak value=6062). The methylation level of zygotes decreased from 2PN stage (peak value=3744) to 4-cell stage (peak value=2826). This methylation level began to rise from 8-cell stage (peak value=3073) to morula stage (peak value=5374), ICM stage (peak value=5706) and TE stage (peak value=8376). The proportion of sperm methylation signal that was in the promoter region was 73.7%, and that in the oocyte was 60.8%, 2PN stage was 57.9%, 4-cell stage was 52.2%, 8-cell stage was 50.3%, morula was 50.3%, ICM was 66.6% and TE was 66.8%. In conclusion, the current study indicated that CpG island methylation changes in human preimplantation embryos were divided into three stages. In the first stage from fertilization to 2PN, the level of CpG island methylation declined sharply. In the second stage from morula to blastular ICM, methylation rapidly increased. In the third stage, methylation was reestablished in the TE. Dynamic CpG island methylation changes were derived primarily from methylation in the promoter region.
Keywords: CpG island methylation; human oocyte; human sperm; preimplantation embryos.
Figures



References
-
- He W, Kang X, Du H, Song B, Lu Z, Huang Y, Wang D, Sun X, Yu Y, Fan Y. Defining differentially methylated regions specific for the acquisition of pluripotency and maintenance in human pluripotent stem cells via microarray. PLoS One. 2014;9:e108350. doi: 10.1371/journal.pone.0108350. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
