Computational analysis of data from a genome-wide screening identifies new PARP1 functional interactors as potential therapeutic targets
- PMID: 31105872
- PMCID: PMC6505629
- DOI: 10.18632/oncotarget.26812
Computational analysis of data from a genome-wide screening identifies new PARP1 functional interactors as potential therapeutic targets
Abstract
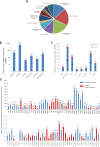
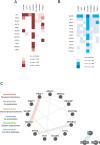
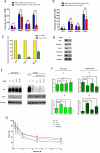
Knowledge of interaction network between different proteins can be a useful tool in cancer therapy. To develop new therapeutic treatments, understanding how these proteins contribute to dysregulated cellular pathways is an important task. PARP1 inhibitors are drugs used in cancer therapy, in particular where DNA repair is defective. It is crucial to find new candidate interactors of PARP1 as new therapeutic targets in order to increase efficacy of PARP1 inhibitors and expand their clinical utility. By a yeast-based genome wide screening, we previously discovered 90 candidate deletion genes that suppress growth-inhibition phenotype conferred by PARP1 in yeast. Here, we performed an integrated and computational analysis to deeply study these genes. First, we identified which pathways these genes are involved in and putative relations with PARP1 through g:Profiler. Then, we studied mutation pattern and their relation to cancer by interrogating COSMIC and DisGeNET database; finally, we evaluated expression and alteration in several cancers with cBioPortal, and the interaction network with GeneMANIA. We identified 12 genes belonging to PARP1-related pathways. We decided to further validate RIT1, INCENP and PSTA1 in MCF7 breast cancer cells. We found that RIT1 and INCENP affected PARylation and PARP1 protein level more significantly in PARP1 inhibited cells. Furthermore, downregulation of RIT1, INCENP and PSAT1 affected olaparib sensitivity of MCF7 cells. Our study identified candidate genes that could have an effect on PARP inhibition therapy. Moreover, we also confirm that yeast-based screenings could be very helpful to identify novel potential therapy factors.
Keywords: PARP1; cancer therapy targets; functional interactors; genome wide screening.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare they have no conflicts of interest.
Figures
Similar articles
-
Expression of human poly (ADP-ribose) polymerase 1 in Saccharomyces cerevisiae: Effect on survival, homologous recombination and identification of genes involved in intracellular localization.Mutat Res. 2015 Apr;774:14-24. doi: 10.1016/j.mrfmmm.2015.02.006. Epub 2015 Mar 6. Mutat Res. 2015. PMID: 25779917
-
RBR-type E3 ubiquitin ligase RNF144A targets PARP1 for ubiquitin-dependent degradation and regulates PARP inhibitor sensitivity in breast cancer cells.Oncotarget. 2017 Oct 10;8(55):94505-94518. doi: 10.18632/oncotarget.21784. eCollection 2017 Nov 7. Oncotarget. 2017. PMID: 29212245 Free PMC article.
-
Novel inhibitors of poly(ADP-ribose) polymerase/PARP1 and PARP2 identified using a cell-based screen in yeast.Cancer Res. 2001 May 15;61(10):4175-83. Cancer Res. 2001. PMID: 11358842
-
Medicinal chemistry approaches of poly ADP-Ribose polymerase 1 (PARP1) inhibitors as anticancer agents - A recent update.Eur J Med Chem. 2019 Mar 1;165:198-215. doi: 10.1016/j.ejmech.2019.01.024. Epub 2019 Jan 12. Eur J Med Chem. 2019. PMID: 30684797 Review.
-
Poly(ADP-Ribose) Polymerase 1: Cellular Pluripotency, Reprogramming, and Tumorogenesis.Int J Mol Sci. 2015 Jul 9;16(7):15531-45. doi: 10.3390/ijms160715531. Int J Mol Sci. 2015. PMID: 26184161 Free PMC article. Review.
Cited by
-
DNA Repair Genes as Drug Candidates for Early Breast Cancer Onset in Latin America: A Systematic Review.Int J Mol Sci. 2021 Dec 2;22(23):13030. doi: 10.3390/ijms222313030. Int J Mol Sci. 2021. PMID: 34884835 Free PMC article.
-
The Genetic Background of Abnormalities in Metabolic Pathways of Phosphoinositides and Their Linkage with the Myotubular Myopathies, Neurodegenerative Disorders, and Carcinogenesis.Biomolecules. 2023 Oct 19;13(10):1550. doi: 10.3390/biom13101550. Biomolecules. 2023. PMID: 37892232 Free PMC article. Review.
-
A Theoretical Study of the Interaction of PARP-1 with Natural and Synthetic Inhibitors: Advances in the Therapy of Triple-Negative Breast Cancer.Curr Issues Mol Biol. 2024 Aug 27;46(9):9415-9429. doi: 10.3390/cimb46090558. Curr Issues Mol Biol. 2024. PMID: 39329910 Free PMC article.
-
Inhibition of DNA Repair in Cancer Therapy: Toward a Multi-Target Approach.Int J Mol Sci. 2020 Sep 12;21(18):6684. doi: 10.3390/ijms21186684. Int J Mol Sci. 2020. PMID: 32932697 Free PMC article. Review.
-
Anticancer drug discovery from Iranian Chrysanthemum cultivars through system pharmacology exploration and experimental validation.Sci Rep. 2021 Jun 3;11(1):11767. doi: 10.1038/s41598-021-91010-y. Sci Rep. 2021. PMID: 34083561 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

