Genome-wide target interactome profiling reveals a novel EEF1A1 epigenetic pathway for oncogenic lncRNA MALAT1 in breast cancer
- PMID: 31105998
- PMCID: PMC6511647
Genome-wide target interactome profiling reveals a novel EEF1A1 epigenetic pathway for oncogenic lncRNA MALAT1 in breast cancer
Erratum in
-
Erratum: Genome-wide target interactome profiling reveals a novel EEF1A1 epigenetic pathway for oncogenic lncRNA MALAT1 in breast cancer.Am J Cancer Res. 2023 Sep 15;13(9):4490-4491. eCollection 2023. Am J Cancer Res. 2023. PMID: 37818059 Free PMC article.
Abstract
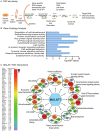
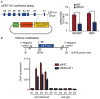
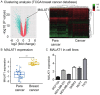
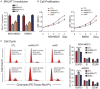
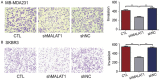
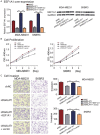
Breast cancer is the most common cancer in women worldwide, accounting for approximately 500,000 deaths each year. MALAT1 is a highly conserved long noncoding RNA (lncRNA), and its increased expression is associated with relapse and metastatic progression in breast cancer. We performed RNA reverse transcription-associated trap sequencing (RAT-seq) to characterize the genome-wide target interaction network for MALAT1 and showed that MALAT1 interacted with multiple pathway target genes that are closely related to tumor progression and metastasis. Notably, MALAT1 bound to the promoter regulatory element of the translation elongation factor 1-alpha 1 gene EEF1A1. Knockdown of MALAT1 by shRNA caused significant downregulation of EEF1A1 in breast cancer MDA-MB231 and SKRB3 cells. Using a luciferase reporter assay, we showed that knockdown of MALAT1 reduced the promoter activity of EEF1A1 in these two breast cancer cells. Chromatin immunoprecipitation (ChIP) assay indicated that MALAT1 regulated EEF1A1 by altering the histone 3 lysine 4 (H3K4) epigenotype in the gene promoter. MALAT1 was overexpressed in breast cancer tissues and breast cancer cells. Knockdown of MALAT1 reduced cell proliferation and invasion by arresting cells at the G0/G1 phase. Ectopic overexpression of EEF1A1 reversed the altered tumor phenotypes induced by MALAT1 shRNA treatment. These data suggest an epigenetic mechanism by which MALAT1 lncRNA facilitates a pro-metastatic phenotype in breast cancer by trans-regulating EEF1A1.
Keywords: EEF1A1; LncRNA; MALAT1; breast cancer; epigenetics; histone methylation; interactome.
Conflict of interest statement
None.
Figures

Similar articles
-
JMJD2C promotes colorectal cancer metastasis via regulating histone methylation of MALAT1 promoter and enhancing β-catenin signaling pathway.J Exp Clin Cancer Res. 2019 Oct 29;38(1):435. doi: 10.1186/s13046-019-1439-x. J Exp Clin Cancer Res. 2019. PMID: 31665047 Free PMC article.
-
Erratum: Genome-wide target interactome profiling reveals a novel EEF1A1 epigenetic pathway for oncogenic lncRNA MALAT1 in breast cancer.Am J Cancer Res. 2023 Sep 15;13(9):4490-4491. eCollection 2023. Am J Cancer Res. 2023. PMID: 37818059 Free PMC article.
-
Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa-miR-448.BMC Cancer. 2016 Feb 25;16:160. doi: 10.1186/s12885-016-2108-5. BMC Cancer. 2016. PMID: 26917489 Free PMC article.
-
Opposing roles of C/EBPα and eEF1A1 in Sp1-regulated miR-122 transcription.RNA Biol. 2020 Feb;17(2):202-210. doi: 10.1080/15476286.2019.1673656. Epub 2019 Oct 7. RNA Biol. 2020. PMID: 31561740 Free PMC article.
-
The Oncogenic and Tumor Suppressive Functions of the Long Noncoding RNA MALAT1: An Emerging Controversy.Front Genet. 2020 Feb 27;11:93. doi: 10.3389/fgene.2020.00093. eCollection 2020. Front Genet. 2020. PMID: 32174966 Free PMC article. Review.
Cited by
-
Downregulation of MALAT1 in triple-negative breast cancer cells.Biochem Biophys Rep. 2023 Nov 28;37:101592. doi: 10.1016/j.bbrep.2023.101592. eCollection 2024 Mar. Biochem Biophys Rep. 2023. PMID: 38088951 Free PMC article.
-
A six-gene signature related with tumor mutation burden for predicting lymph node metastasis in breast cancer.Transl Cancer Res. 2021 May;10(5):2229-2246. doi: 10.21037/tcr-20-3471. Transl Cancer Res. 2021. PMID: 35116541 Free PMC article.
-
Long Non-Coding RNA: Dual Effects on Breast Cancer Metastasis and Clinical Applications.Cancers (Basel). 2019 Nov 16;11(11):1802. doi: 10.3390/cancers11111802. Cancers (Basel). 2019. PMID: 31744046 Free PMC article. Review.
-
Potential roles of lncRNA MALAT1-miRNA interactions in ocular diseases.J Cell Commun Signal. 2023 Dec;17(4):1203-1217. doi: 10.1007/s12079-023-00787-2. Epub 2023 Oct 23. J Cell Commun Signal. 2023. PMID: 37870615 Free PMC article. Review.
-
Long Noncoding RNAs Involved in the Endocrine Therapy Resistance of Breast Cancer.Cancers (Basel). 2020 May 31;12(6):1424. doi: 10.3390/cancers12061424. Cancers (Basel). 2020. PMID: 32486413 Free PMC article. Review.
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. - PubMed
-
- Hosono Y, Niknafs YS, Prensner JR, Iyer MK, Dhanasekaran SM, Mehra R, Pitchiaya S, Tien J, Escara-Wilke J, Poliakov A, Chu SC, Saleh S, Sankar K, Su F, Guo S, Qiao Y, Freier SM, Bui HH, Cao X, Malik R, Johnson TM, Beer DG, Feng FY, Zhou W, Chinnaiyan AM. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell. 2017;171:1559–1572. e20. - PMC - PubMed
-
- Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J, Rao X, Li M, Sun M, Jiang M, Xu Y, Teng L, Jin Y, Si J, Liu W, Kang Y, Zhou T. Long non-coding RNA GMAN, upregulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of ephrin A1 by competitively binding GMAN-AS. Gastroenterology. 2019;156:676–691. e11. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
