The Role of Structural Polymorphism in Driving the Mechanical Performance of the Alzheimer's Beta Amyloid Fibrils
- PMID: 31106199
- PMCID: PMC6499180
- DOI: 10.3389/fbioe.2019.00083
The Role of Structural Polymorphism in Driving the Mechanical Performance of the Alzheimer's Beta Amyloid Fibrils
Abstract
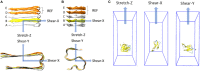
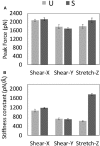
Alzheimer's Disease (AD) is related with the abnormal aggregation of amyloid β-peptides Aβ1-40 and Aβ1-42, the latter having a polymorphic character which gives rise to U- or S-shaped fibrils. Elucidating the role played by the nanoscale-material architecture on the amyloid fibril stability is a crucial breakthrough to better understand the pathological nature of amyloid structures and to support the rational design of bio-inspired materials. The computational study here presented highlights the superior mechanical behavior of the S-architecture, characterized by a Young's modulus markedly higher than the U-shaped architecture. The S-architecture showed a higher mechanical resistance to the enforced deformation along the fibril axis, consequence of a better interchain hydrogen bonds' distribution. In conclusion, this study, focusing the attention on the pivotal multiscale relationship between molecular phenomena and material properties, suggests the S-shaped Aβ1-42 species as a target of election in computational screen/design/optimization of effective aggregation modulators.
Keywords: Alzheimer's Disease; Young Modulus; amyloid fibrils; biomechanics; molecular dynamics simulations; structural polymorphism.
Figures


Similar articles
-
Conformational Dynamics and Stability of U-Shaped and S-Shaped Amyloid β Assemblies.Int J Mol Sci. 2018 Feb 14;19(2):571. doi: 10.3390/ijms19020571. Int J Mol Sci. 2018. PMID: 29443891 Free PMC article.
-
Mutations alter the geometry and mechanical properties of Alzheimer's Aβ(1-40) amyloid fibrils.Biochemistry. 2010 Oct 19;49(41):8967-77. doi: 10.1021/bi100953t. Biochemistry. 2010. PMID: 20731379
-
S14G-humanin inhibits Aβ1-42 fibril formation, disaggregates preformed fibrils, and protects against Aβ-induced cytotoxicity in vitro.J Pept Sci. 2013 Mar;19(3):159-65. doi: 10.1002/psc.2484. Epub 2013 Jan 24. J Pept Sci. 2013. Retraction in: J Pept Sci. 2016 Jun;22(6):434. doi: 10.1002/psc.2889. PMID: 23349038 Retracted.
-
Recent Advances by In Silico and In Vitro Studies of Amyloid-β 1-42 Fibril Depicted a S-Shape Conformation.Int J Mol Sci. 2018 Aug 16;19(8):2415. doi: 10.3390/ijms19082415. Int J Mol Sci. 2018. PMID: 30115846 Free PMC article. Review.
-
Elucidating the Structures of Amyloid Oligomers with Macrocyclic β-Hairpin Peptides: Insights into Alzheimer's Disease and Other Amyloid Diseases.Acc Chem Res. 2018 Mar 20;51(3):706-718. doi: 10.1021/acs.accounts.7b00554. Epub 2018 Mar 6. Acc Chem Res. 2018. PMID: 29508987 Free PMC article. Review.
Cited by
-
When Stiffness Matters: Mechanosensing in Heart Development and Disease.Front Cell Dev Biol. 2020 May 25;8:334. doi: 10.3389/fcell.2020.00334. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32671058 Free PMC article. Review.
-
Inhibitory Activity of Insulin on Aβ Aggregation Is Restricted Due to Binding Selectivity and Specificity to Polymorphic Aβ States.ACS Chem Neurosci. 2020 Feb 5;11(3):445-452. doi: 10.1021/acschemneuro.9b00645. Epub 2020 Jan 10. ACS Chem Neurosci. 2020. PMID: 31899862 Free PMC article.
-
Coarse-grained MD simulations reveal beta-amyloid fibrils of various sizes bind to interfacial liquid-ordered and liquid-disordered regions in phase separated lipid rafts with diverse membrane-bound conformational states.Biophys Chem. 2020 May;260:106355. doi: 10.1016/j.bpc.2020.106355. Epub 2020 Mar 5. Biophys Chem. 2020. PMID: 32179374 Free PMC article.
-
The Impact of Natural Compounds on S-Shaped Aβ42 Fibril: From Molecular Docking to Biophysical Characterization.Int J Mol Sci. 2020 Mar 16;21(6):2017. doi: 10.3390/ijms21062017. Int J Mol Sci. 2020. PMID: 32188076 Free PMC article.
-
Destabilization of the Alzheimer's amyloid-β peptide by a proline-rich β-sheet breaker peptide: a molecular dynamics simulation study.J Mol Model. 2021 Nov 18;27(12):356. doi: 10.1007/s00894-021-04968-x. J Mol Model. 2021. PMID: 34796404
References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Adamcik J., Berquand A., Mezzenga R. (2011). Single-step direct measurement of amyloid fibrils stiffness by peak force quantitative nanomechanical atomic force microscopy. Appl. Phys. Lett. 98, 1–4. 10.1063/1.3589369 - DOI
-
- Adamcik J., Mezzenga R. (2012). Study of amyloid fibrils via atomic force microscopy. Curr. Opin. Colloid Interface Sci. 17, 369–376. 10.1016/j.cocis.2012.08.001 - DOI
LinkOut - more resources
Full Text Sources

