Rapid functional activation of a horizontally transferred eukaryotic gene in a bacterial genome in the absence of selection
- PMID: 31106341
- PMCID: PMC6614815
- DOI: 10.1093/nar/gkz370
Rapid functional activation of a horizontally transferred eukaryotic gene in a bacterial genome in the absence of selection
Abstract
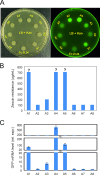
Horizontal gene transfer has occurred between organisms of all domains of life and contributed substantially to genome evolution in both prokaryotes and eukaryotes. Phylogenetic evidence suggests that eukaryotic genes horizontally transferred to bacteria provided useful new gene functions that improved metabolic plasticity and facilitated adaptation to new environments. How these eukaryotic genes evolved into functional bacterial genes is not known. Here, we have conducted a genetic screen to identify the mechanisms involved in functional activation of a eukaryotic gene after its transfer into a bacterial genome. We integrated a eukaryotic selectable marker gene cassette driven by expression elements from the red alga Porphyridium purpureum into the genome of Escherichia coli. Following growth under non-selective conditions, gene activation events were indentified by antibiotic selection. We show that gene activation in the bacterial recipient occurs at high frequency and involves two major types of spontaneous mutations: deletion and gene amplification. We further show that both mechanisms result in promoter capture and are frequently triggered by microhomology-mediated recombination. Our data suggest that horizontally transferred genes have a high probability of acquiring functionality, resulting in their maintenance if they confer a selective advantage.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Tandem gene duplication selected by activation of horizontally transferred gene in bacteria.Appl Microbiol Biotechnol. 2024 May 23;108(1):340. doi: 10.1007/s00253-024-13160-z. Appl Microbiol Biotechnol. 2024. PMID: 38777914 Free PMC article.
-
Rapid functional activation of horizontally transferred eukaryotic intron-containing genes in the bacterial recipient.Nucleic Acids Res. 2024 Aug 12;52(14):8344-8355. doi: 10.1093/nar/gkae628. Nucleic Acids Res. 2024. PMID: 39011898 Free PMC article.
-
Replication of bacterial plasmids in the nucleus of the red alga Porphyridium purpureum.Nat Commun. 2018 Aug 27;9(1):3451. doi: 10.1038/s41467-018-05651-1. Nat Commun. 2018. PMID: 30150628 Free PMC article.
-
Horizontal gene transfer in prokaryotes: quantification and classification.Annu Rev Microbiol. 2001;55:709-42. doi: 10.1146/annurev.micro.55.1.709. Annu Rev Microbiol. 2001. PMID: 11544372 Free PMC article. Review.
-
Functional horizontal gene transfer from bacteria to eukaryotes.Nat Rev Microbiol. 2018 Feb;16(2):67-79. doi: 10.1038/nrmicro.2017.137. Epub 2017 Nov 27. Nat Rev Microbiol. 2018. PMID: 29176581 Review.
Cited by
-
A hidden intrinsic ability of bicistronic expression based on a novel translation reinitiation mechanism in yeast.Nucleic Acids Res. 2025 Mar 20;53(6):gkaf220. doi: 10.1093/nar/gkaf220. Nucleic Acids Res. 2025. PMID: 40156854 Free PMC article.
-
Optimized transgene expression in the red alga Porphyridium purpureum and efficient recombinant protein secretion into the culture medium.Plant Mol Biol. 2024 Feb 14;114(1):18. doi: 10.1007/s11103-024-01415-2. Plant Mol Biol. 2024. PMID: 38353826 Free PMC article.
-
Natural bacterial isolates as an inexhaustible source of new bacteriocins.Appl Microbiol Biotechnol. 2021 Jan;105(2):477-492. doi: 10.1007/s00253-020-11063-3. Epub 2021 Jan 4. Appl Microbiol Biotechnol. 2021. PMID: 33394148 Review.
-
Tandem gene duplication selected by activation of horizontally transferred gene in bacteria.Appl Microbiol Biotechnol. 2024 May 23;108(1):340. doi: 10.1007/s00253-024-13160-z. Appl Microbiol Biotechnol. 2024. PMID: 38777914 Free PMC article.
-
Rapid functional activation of horizontally transferred eukaryotic intron-containing genes in the bacterial recipient.Nucleic Acids Res. 2024 Aug 12;52(14):8344-8355. doi: 10.1093/nar/gkae628. Nucleic Acids Res. 2024. PMID: 39011898 Free PMC article.
References
-
- Vaughn J.C., Mason M.T., Sper-Whitis G.L., Kuhlman P., Palmer J.D.. Fungal origin by horizontal gene transfer of a plant mitochondrial group I intron in the chimeric coxI gene of Peperomia. J. Mol. Evol. 1995; 41:563–572. - PubMed
-
- Bergthorsson U., Adams K.L., Thomason B., Palmer J.D.. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003; 424:197–201. - PubMed
-
- Denker E., Bapteste E., Le Guyader H., Manuel M., Rabet N.. Horizontal gene transfer and the evolution of cnidarian stinging cells. Curr. Biol. 2008; 18:R858–R859. - PubMed
-
- Gladyshev E.A., Meselson M., Arkhipova I.R.. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008; 320:1210–1213. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

