NAPS update: network analysis of molecular dynamics data and protein-nucleic acid complexes
- PMID: 31106363
- PMCID: PMC6602509
- DOI: 10.1093/nar/gkz399
NAPS update: network analysis of molecular dynamics data and protein-nucleic acid complexes
Abstract
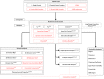
Network theory is now a method of choice to gain insights in understanding protein structure, folding and function. In combination with molecular dynamics (MD) simulations, it is an invaluable tool with widespread applications such as analyzing subtle conformational changes and flexibility regions in proteins, dynamic correlation analysis across distant regions for allosteric communications, in drug design to reveal alternative binding pockets for drugs, etc. Updated version of NAPS now facilitates network analysis of the complete repertoire of these biomolecules, i.e., proteins, protein-protein/nucleic acid complexes, MD trajectories, and RNA. Various options provided for analysis of MD trajectories include individual network construction and analysis of intermediate time-steps, comparative analysis of these networks, construction and analysis of average network of the ensemble of trajectories and dynamic cross-correlations. For protein-nucleic acid complexes, networks of the whole complex as well as that of the interface can be constructed and analyzed. For analysis of proteins, protein-protein complexes and MD trajectories, network construction based on inter-residue interaction energies with realistic edge-weights obtained from standard force fields is provided to capture the atomistic details. Updated version of NAPS also provides improved visualization features, interactive plots and bulk execution. URL: http://bioinf.iiit.ac.in/NAPS/.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

