Genome-wide identification of GhAAI genes reveals that GhAAI66 triggers a phase transition to induce early flowering
- PMID: 31106831
- PMCID: PMC6760319
- DOI: 10.1093/jxb/erz239
Genome-wide identification of GhAAI genes reveals that GhAAI66 triggers a phase transition to induce early flowering
Abstract
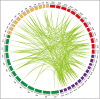
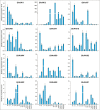
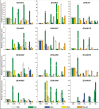
Plants undergo a phase transition from vegetative to reproductive development that triggers floral induction. Genes containing an AAI (α-amylase inhibitor) domain form a large gene family, but there have been no comprehensive analyses of this gene family in any plant species. Here, we identified 336 AAI genes from nine plant species including122 AAI genes in cotton (Gossypium hirsutum). The AAI gene family has evolutionarily conserved amino acid residues throughout the plant kingdom. Phylogenetic analysis classified AAI genes into five major clades with significant polyploidization and showing effects of genome duplication. Our study identified 42 paralogous and 216 orthologous gene pairs resulting from segmental and whole-genome duplication, respectively, demonstrating significant contributions of gene duplication to expansion of the cotton AAI gene family. Further, GhAAI66 was preferentially expressed in flower tissue and as responses to phytohormone treatments. Ectopic expression of GhAAI66 in Arabidopsis and silencing in cotton revealed that GhAAI66 triggers a phase transition to induce early flowering. Further, GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis of RNA sequencing data and qRT-PCR (quantitative reverse transcription-PCR) analysis indicated that GhAAI66 integrates multiple flower signaling pathways including gibberellin, jasmonic acid, and floral integrators to trigger an early flowering cascade in Arabidopsis. Therefore, characterization of the AAI family provides invaluable insights for improving cotton breeding.
Keywords: Gossypium hirsutum; Early flowering; ectopic expression; floral integrators; gene duplication; phylogenetic analysis.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures




References
-
- Bao Y, Hu GJ, Flagel LE, Salmon A, Bezanilla M, Paterson AH, Wang ZN, Wendel JF. 2011. Parallel up-regulation of the profilin gene family following independent domestication of diploid and allopolyploid cotton (Gossypium). Proceedings of the National Academy of Sciences, USA 108, 21152–21157. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

