Nuclear retention element recruits U1 snRNP components to restrain spliced lncRNAs in the nucleus
- PMID: 31107149
- PMCID: PMC6602561
- DOI: 10.1080/15476286.2019.1620061
Nuclear retention element recruits U1 snRNP components to restrain spliced lncRNAs in the nucleus
Abstract
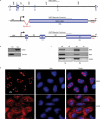
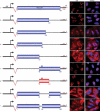
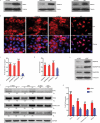
In contrast to cytoplasmic localization of spliced mRNAs, many spliced lncRNAs are localized in the nucleus. To investigate the mechanism, we used lncRNA MEG3 as a reporter and mapped a potent nuclear retention element (NRE), deletion of this element led to striking export of MEG3 from the nucleus to the cytoplasm. Insertion of the NRE resulted in nuclear retention of spliced lncRNA as well as spliced mRNA. We further purified RNP assembled on the NRE in vitro and identified the proteins by mass spectrometry. Screen using siRNA revealed depletion of U1 snRNP components SNRPA, SNRNP70 or SNRPD2 caused significant cytoplasmic localization of MEG3 reporter transcripts. Co-knockdown these factors in HFF1 cells resulted in an increased cytoplasmic distribution of endogenous lncRNAs. Together, these data support a model that U1 snRNP components restrain spliced lncRNAs in the nucleus via the interaction with nuclear retention element.
Keywords: LncRNA localization; MEG3; U1 snRNP components; nuclear retention element.
Figures

References
-
- Cheng H, Dufu K, Lee CS, et al. Human mRNA export machinery recruited to the 5ʹ end of mRNA. Cell. 2006;127:1389–1400. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
