Coalescing replication compartments provide the opportunity for recombination between coinfecting herpesviruses
- PMID: 31107607
- PMCID: PMC6662979
- DOI: 10.1096/fj.201900032R
Coalescing replication compartments provide the opportunity for recombination between coinfecting herpesviruses
Erratum in
-
Erratum to "Coalescing replication compartments provide the opportunity for recombination between coinfecting herpesviruses".FASEB J. 2022 Oct;36(10):e22581. doi: 10.1096/fsb2.22581. FASEB J. 2022. PMID: 36169184 Free PMC article. No abstract available.
Abstract
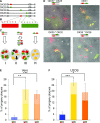
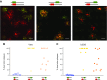
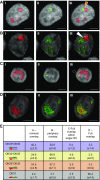
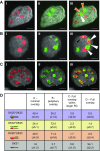
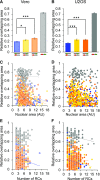
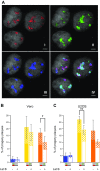
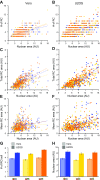
Homologous recombination (HR) is considered a major driving force of evolution because it generates and expands genetic diversity. Evidence of HR between coinfecting herpesvirus DNA genomes can be found frequently both in vitro and in clinical isolates. Each herpes simplex virus type 1 (HSV-1) replication compartment (RC) derives from a single incoming genome and maintains a specific territory within the nucleus. This raises intriguing questions about where and when coinfecting viral genomes interact. To study the spatiotemporal requirements for intergenomic recombination, we developed an assay with dual-color FISH that enables detection of HR between different pairs of coinfecting HSV-1 genomes. Our results revealed that HR increases intermingling of RCs derived from different genomes. Furthermore, inhibition of RC movement reduces the rate of HR events among coinfecting viruses. Finally, we observed correlation between nuclear size and the number of RCs per nucleus. Our findings suggest that both viral replication and recombination are subject to nuclear spatial constraints. Other DNA viruses and cellular DNA are likely to encounter similar restrictions.-Tomer, E., Cohen, E. M., Drayman, N., Afriat, A., Weitzman, M. D., Zaritsky, A., Kobiler, O. Coalescing replication compartments provide the opportunity for recombination between coinfecting herpesviruses.
Keywords: DNA recombination; herpes simplex virus; nuclear spatial constraints; viral nuclear interactions.
Conflict of interest statement
The authors thank Eitan Erez Zahavi, Ariel Ionescu, and Eran Perlson for advice and valuable support and all Kobiler lab members for their comments. This work was supported by grants from the Israel Science Foundation (Grant 1387/14) to O.K., the European Union Career Integration Grant (EU CIG) (FP7-2012-333653) to O.K., the U.S. National Institutes of Health (NIH), National Institute of Neurological disorders (NS082240) to M.D.W., the United States - Israel Binational Science Foundation (Grant 2015395) to O.K. and M.D.W., a Marguerite Stolz Research Fellowship to O.K. and E.T. is supported by the Buchman Scholarship, and A.Z. was supported by a Grant from the NIH, National Institute of General Medical Sciences (P01 GM103723) to Gaudenz Danuser (Departments of Bioinformatics and Cell Biology, University of Texas Southwestern Medical Center, Dallas, TX). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors declare no conflicts of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

