Triggering NETosis via protease-activated receptor (PAR)-2 signaling as a mechanism of hijacking neutrophils function for pathogen benefits
- PMID: 31107907
- PMCID: PMC6544335
- DOI: 10.1371/journal.ppat.1007773
Triggering NETosis via protease-activated receptor (PAR)-2 signaling as a mechanism of hijacking neutrophils function for pathogen benefits
Abstract
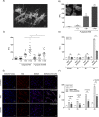
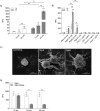
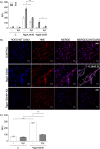
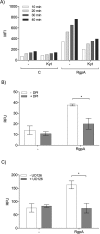
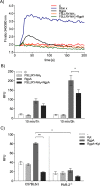
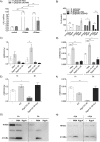
Neutrophil-derived networks of DNA-composed extracellular fibers covered with antimicrobial molecules, referred to as neutrophil extracellular traps (NETs), are recognized as a physiological microbicidal mechanism of innate immunity. The formation of NETs is also classified as a model of a cell death called NETosis. Despite intensive research on the NETs formation in response to pathogens, the role of specific bacteria-derived virulence factors in this process, although postulated, is still poorly understood. The aim of our study was to determine the role of gingipains, cysteine proteases responsible for the virulence of P. gingivalis, on the NETosis process induced by this major periodontopathogen. We showed that NETosis triggered by P. gingivalis is gingipain dependent since in the stark contrast to the wild-type strain (W83) the gingipain-null mutant strain only slightly induced the NETs formation. Furthermore, the direct effect of proteases on NETosis was documented using purified gingipains. Notably, the induction of NETosis was dependent on the catalytic activity of gingipains, since proteolytically inactive forms of enzymes showed reduced ability to trigger the NETs formation. Mechanistically, gingipain-induced NETosis was dependent on proteolytic activation of protease-activated receptor-2 (PAR-2). Intriguingly, both P. gingivalis and purified Arg-specific gingipains (Rgp) induced NETs that not only lacked bactericidal activity but instead stimulated the growth of bacteria species otherwise susceptible to killing in NETs. This protection was executed by proteolysis of bactericidal components of NETs. Taken together, gingipains play a dual role in NETosis: they are the potent direct inducers of NETs formation but in the same time, their activity prevents P. gingivalis entrapment and subsequent killing. This may explain a paradox that despite the massive accumulation of neutrophils and NETs formation in periodontal pockets periodontal pathogens and associated pathobionts thrive in this environment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
The role of phagocytosis, oxidative burst and neutrophil extracellular traps in the interaction between neutrophils and the periodontal pathogen Porphyromonas gingivalis.Mol Oral Microbiol. 2015 Oct;30(5):361-75. doi: 10.1111/omi.12099. Epub 2015 May 4. Mol Oral Microbiol. 2015. PMID: 25869817
-
Gingipains: Critical Factors in the Development of Aspiration Pneumonia Caused by Porphyromonas gingivalis.J Innate Immun. 2016;8(2):185-98. doi: 10.1159/000441724. Epub 2015 Nov 28. J Innate Immun. 2016. PMID: 26613585 Free PMC article.
-
Gingipains protect Porphyromonas gingivalis from macrophage-mediated phagocytic clearance.PLoS Pathog. 2025 Jan 21;21(1):e1012821. doi: 10.1371/journal.ppat.1012821. eCollection 2025 Jan. PLoS Pathog. 2025. PMID: 39836688 Free PMC article.
-
Role of gingipains R in the pathogenesis of Porphyromonas gingivalis-mediated periodontal disease.Clin Infect Dis. 1999 Mar;28(3):456-65. doi: 10.1086/515156. Clin Infect Dis. 1999. PMID: 10194062 Review.
-
Manipulation of Neutrophils by Porphyromonas gingivalis in the Development of Periodontitis.Front Cell Infect Microbiol. 2017 May 23;7:197. doi: 10.3389/fcimb.2017.00197. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28589098 Free PMC article. Review.
Cited by
-
Antibodies to Porphyromonas gingivalis Are Increased in Patients with Severe Periodontitis, and Associate with Presence of Specific Autoantibodies and Myocardial Infarction.J Clin Med. 2022 Feb 15;11(4):1008. doi: 10.3390/jcm11041008. J Clin Med. 2022. PMID: 35207282 Free PMC article.
-
Neutrophils' Contribution to Periodontitis and Periodontitis-Associated Cardiovascular Diseases.Int J Mol Sci. 2023 Oct 19;24(20):15370. doi: 10.3390/ijms242015370. Int J Mol Sci. 2023. PMID: 37895050 Free PMC article. Review.
-
Subversion of Lipopolysaccharide Signaling in Gingival Keratinocytes via MCPIP-1 Degradation as a Novel Pathogenic Strategy of Inflammophilic Pathobionts.mBio. 2021 Jun 29;12(3):e0050221. doi: 10.1128/mBio.00502-21. Epub 2021 Jun 29. mBio. 2021. PMID: 34182783 Free PMC article.
-
Non-classical neutrophil extracellular traps induced by PAR2-signaling proteases.Cell Death Dis. 2025 Feb 19;16(1):109. doi: 10.1038/s41419-025-07428-z. Cell Death Dis. 2025. PMID: 39971938 Free PMC article.
-
Porphyromonas gingivalis affects neutrophil pro-inflammatory activities.Front Cell Dev Biol. 2025 Jan 28;13:1419651. doi: 10.3389/fcell.2025.1419651. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 39936030 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

