Tyrosines involved in the activity of φ29 single-stranded DNA binding protein
- PMID: 31107918
- PMCID: PMC6527236
- DOI: 10.1371/journal.pone.0217248
Tyrosines involved in the activity of φ29 single-stranded DNA binding protein
Abstract
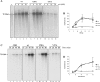
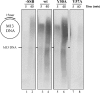
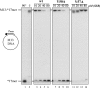
The genome of Bacillus subtilis phage ϕ29 consists of a linear double-stranded DNA with a terminal protein (TP) covalently linked to each 5' end (TP-DNA). ϕ29 DNA polymerase is the enzyme responsible for viral DNA replication, due to its distinctive properties: high processivity and strand displacement capacity, being able to replicate the entire genome without requiring the assistance of processivity or unwinding factors, unlike most replicases. ϕ29 single-stranded DNA binding protein (SSB) is encoded by the viral gene 5 and binds the ssDNA generated in the replication of the ϕ29 TP-DNA. It has been described to stimulate the DNA elongation rate during the DNA replication. Previous studies proposed residues Tyr50, Tyr57 and Tyr76 as ligands of ssDNA. The role of two of these residues has been determined in this work by site-directed mutagenesis. Our results showed that mutant derivative Y57A was unable to bind to ssDNA, to stimulate the DNA elongation and to displace oligonucleotides annealed to M13 ssDNA, whereas mutant Y50A behaved like the wild-type SSB.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Strand Displacement and Unwinding Assays to Study the Concerted Action of the DNA Polymerase and SSB During Phi29 TP-DNA Replication.Methods Mol Biol. 2021;2281:333-342. doi: 10.1007/978-1-0716-1290-3_22. Methods Mol Biol. 2021. PMID: 33847970
-
Helix-destabilizing activity of phi 29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication.J Mol Biol. 1995 Nov 3;253(4):517-29. doi: 10.1006/jmbi.1995.0570. J Mol Biol. 1995. PMID: 7473731
-
Differential functional behavior of viral phi29, Nf and GA-1 SSB proteins.Nucleic Acids Res. 2000 May 15;28(10):2034-42. doi: 10.1093/nar/28.10.2034. Nucleic Acids Res. 2000. PMID: 10773070 Free PMC article.
-
Protein-Primed Replication of Bacteriophage Φ29 DNA.Enzymes. 2016;39:137-67. doi: 10.1016/bs.enz.2016.03.005. Epub 2016 May 12. Enzymes. 2016. PMID: 27241929 Review.
-
Protein-nucleic acid interactions in bacteriophage phi 29 DNA replication.FEMS Microbiol Rev. 1995 Aug;17(1-2):73-82. doi: 10.1111/j.1574-6976.1995.tb00189.x. FEMS Microbiol Rev. 1995. PMID: 7669351 Review.
Cited by
-
Unlimited Cooperativity of Betatectivirus SSB, a Novel DNA Binding Protein Related to an Atypical Group of SSBs From Protein-Primed Replicating Bacterial Viruses.Front Microbiol. 2021 Jun 29;12:699140. doi: 10.3389/fmicb.2021.699140. eCollection 2021. Front Microbiol. 2021. PMID: 34267740 Free PMC article.
-
Strand Displacement and Unwinding Assays to Study the Concerted Action of the DNA Polymerase and SSB During Phi29 TP-DNA Replication.Methods Mol Biol. 2021;2281:333-342. doi: 10.1007/978-1-0716-1290-3_22. Methods Mol Biol. 2021. PMID: 33847970
References
-
- Gutiérrez J, García JA, Blanco L, Salas M. Cloning and template activity of the origins of replication of phage ϕ29 DNA. Gene. 1986;43(1–2):1–11. . - PubMed
-
- Serrano M, Gutiérrez C, Freire R, Bravo A, Salas M, Hermoso JM. Phage ϕ29 protein p6: a viral histone-like protein. Biochimie. 1994;76(10–11):981–91. . - PubMed

