Rice Galaxy: an open resource for plant science
- PMID: 31107941
- PMCID: PMC6527052
- DOI: 10.1093/gigascience/giz028
Rice Galaxy: an open resource for plant science
Erratum in
-
Corrigendum to: Rice Galaxy: an open resource for plant science.Gigascience. 2019 Dec 1;8(12):giz156. doi: 10.1093/gigascience/giz156. Gigascience. 2019. PMID: 31886874 Free PMC article. No abstract available.
Abstract
Background: Rice molecular genetics, breeding, genetic diversity, and allied research (such as rice-pathogen interaction) have adopted sequencing technologies and high-density genotyping platforms for genome variation analysis and gene discovery. Germplasm collections representing rice diversity, improved varieties, and elite breeding materials are accessible through rice gene banks for use in research and breeding, with many having genome sequences and high-density genotype data available. Combining phenotypic and genotypic information on these accessions enables genome-wide association analysis, which is driving quantitative trait loci discovery and molecular marker development. Comparative sequence analyses across quantitative trait loci regions facilitate the discovery of novel alleles. Analyses involving DNA sequences and large genotyping matrices for thousands of samples, however, pose a challenge to non-computer savvy rice researchers.
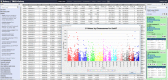
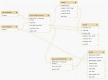
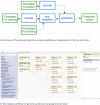
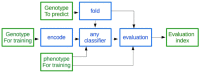
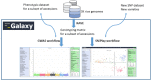
Findings: The Rice Galaxy resource has shared datasets that include high-density genotypes from the 3,000 Rice Genomes project and sequences with corresponding annotations from 9 published rice genomes. The Rice Galaxy web server and deployment installer includes tools for designing single-nucleotide polymorphism assays, analyzing genome-wide association studies, population diversity, rice-bacterial pathogen diagnostics, and a suite of published genomic prediction methods. A prototype Rice Galaxy compliant to Open Access, Open Data, and Findable, Accessible, Interoperable, and Reproducible principles is also presented.
Conclusions: Rice Galaxy is a freely available resource that empowers the plant research community to perform state-of-the-art analyses and utilize publicly available big datasets for both fundamental and applied science.
Keywords: Galaxy project; breeding; genome-wide association studies; genomes; high-density genotypes; reproducibility; rice; single-nucleotide polymorphism; workflow.
© The Author(s) 2019. Published by Oxford University Press.
Figures



References
-
- Mansueto L, Fuentes RR, Chebotarov D, et al.. SNP-Seek II: A resource for allele mining and analysis of big genomic data in Oryza sativa. Curr Plant Biol. 2016;6628:16–25.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

